
A combination of transcriptome and methylation analyses reveals the role of lncRNA HOTAIRM1 in the proliferation and metastasis of breast cancer
Introduction
The incidence of breast invasive carcinoma (BRCA), the most frequently occurring type of carcinoma among women (1,2), is currently on the rise worldwide (3). Despite the improvements in BRCA therapies over the last decade, metastasis, tumor relapse, and resistance to therapy remain the chief causes of BRCA-related deaths in patients (2,4-6). Understanding the specific molecular mechanisms underlying BRCA development and progression is imperative to the effective management of the disease. DNA methylation, an epigenetic modification process catalyzed by DNA methyltransferases such as DNMT1, DNMT3A, and DNMT3B, can regulate gene expression without changing the DNA sequence (7). Previous studies have shown that DNA methylation predominantly occurs in cytosine-phosphate-guanine (CpG) dinucleotides in mammals, accounting for 70–80% of the entire human genome (8-12). Additional evidence has demonstrated that aberrant DNA methylation regulates abnormal gene expression and malignant phenotypes (13,14), while gene expression levels are negatively correlated with DNA methylation factors, named methylation-driven genes (15). In fact, widespread DNA methylation alterations in normal breast tissue adjacent to cancer that become enriched with breast cancer progression have been identified, suggesting that DNA methylation alterations predate the emergence of breast cancer. Furthermore, DNA methylation status has been found to be strongly associated with the prognosis of BRCA patients (16-18). Previous studies demonstrated that silencing genes such as PENl, BCSGl, PLAU, IGF, and CDN3 in human breast cancer cells was associated with methylation status (19,20). These genes are essential in regulating various processes, including cell proliferation, apoptosis, cell cycle progression, and metastasis (21). Although some progress has been made in the development of therapies against BRCA, there is currently no cause for optimism, mainly owing to the lack of clarity regarding the cause of the disease. Therefore, elucidating the mechanisms underlying DNA methylation coupled with understanding its biological function is imperative to guiding the development of effective treatment and prevention therapies for BRCA patients.
In recent decades, researchers have identified and elucidated the biological functions of long noncoding RNAs (lncRNAs) in various human cancers (22,23). Consequently, much attention has been paid to their functions in tumors, including cell proliferation, differentiation, chromosomal remodeling, epigenetic regulation, and transcription and post-transcriptional modification (24,25). For example, recent evidence has demonstrated that H19 is more abundant in estrogen receptor (ER)-positive compared with ER-negative breast tumor tissues, where it promotes tamoxifen resistance (26,27).
In the present study, we combined transcriptome and methylation analyses to reveal an aberrant lncRNA, named HOX antisense intergenic RNA myeloid 1 (HOTAIRM1), in BRCA tissues. This lncRNA not only acts as a tumor suppressor in various human cancers, but can also regulate papillary thyroid cancer cell proliferation and invasion through the HOTAIRM1/miR-107/TDG axis (28). In addition, Kim et al. (29) found that HOXA1 and its neighboring HOTAIRM1 might serve as potential therapeutic targets for ER+ breast cancer patients. To date, however, nothing is known regarding the relationship between DNA methylation of the HOTAIRM1 gene in BRCA. Therefore, additional studies are needed to clarify the role of DNA methylation in regulating HOTAIRM1 expression. For the first time we found DNA methylation is associated with HOTAIRM1 using transcriptome and methylation combined analysis in breast cancer, and regulated by DNMT1 and DNMT3A. Results of the present study demonstrated that HOTAIRM1 is downregulated and hypermethylated in BRCA patients, and plays a critical role in the proliferation and metastasis of breast cancer cells. Notably, this downregulation is a significant prognostic factor for the survival of BRCA patients, and thus may be a promising therapeutic target. We present the following article in accordance with the MDAR reporting checklist (available at https://gs.amegroups.com/article/view/10.21037/gs-22-164/rc).
Methods
Data retrieval
RNA sequencing data, comprising 1,109 BRCA tumor and 113 normal tissues, were retrieved from The Cancer Genome Atlas (TCGA) database (https://tcga-data.nci.nih.gov/tcga). DNA methylation profiles, including 315 BRCA tumor tissues alongside 27 normal tissues, were also downloaded from the TCGA DNA methylation database (Illumina human methylation 27 platform).
Identification of differentially expressed and aberrantly methylated genes
Firstly, we employed the “limma” package, implemented in R software, to identify differentially expressed and aberrantly methylated genes based on the following criteria: |log2fold change (FC)| >1, false discovery rate (FDR) <0.05. Next, we used the “MethylMix” package (30) to screen for upregulated and downregulated hypermethylated genes, with the promoter of these genes generally considered a 2,000 bp sequence upstream of the transcription start site (31). Finally, the same package was used to analyze the distribution of the methylation-driven gene promoters.
Determination of the prognostic value
To explore the prognostic value of the methylation-driven genes, we combined gene methylation and expression data with the survival status and time data of patients, then subjected them to univariate and multivariate analyses using Cox proportional hazards regression models. Multivariate analyses were conducted to determine the prognostic value of the methylation-driven genes and clinical characteristics.
Clinical specimens and approval
We collected 51 breast cancer and 51 corresponding normal adjacent tissues from patients at the First Affiliated Hospital of Gannan Medical University, China. The samples were immediately frozen in liquid nitrogen following surgical resection. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The present study was approved by the Ethics Committee of the First Affiliated Hospital of Gannan Medical University (No. LLSC-2022033101). Informed consent was obtained from patients or guardians.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from the collected breast cancer and normal tissues and cells using an RNA isolation kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. The RNA was reverse transcribed into complementary DNA (cDNA) using a reverse transcriptase kit (TaKaRa, Dalian, China). The cDNA was subjected to qRT-PCR using the Takara SYBR® Premix Ex TaqTM II kit (TaKaRa, Dalian, China), and qRT-PCR was performed on a 7500 Real-Time PCR System (ABI) targeting genes whose primer sequences are listed in Table S1.
Cell cultures and lentiviral transfection
Human breast cancer cell lines, MCF7 and T47D, were purchased from the Chinese Academy of Sciences Cell Bank (Shanghai, China) and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Corning, VA, USA) supplemented with 10% fetal bovine serum (FBS; Corning, VA, USA).
Constructs for overexpression of HOTAIRM1, DNMT1, and DNMT3B (LV-HOTAIRM1, LV-DNMT1, and LV-DNMT3B) were constructed and transfected in MCF7 and T47D cells using lentiviral overexpression plasmids Ubi-MCS-SV40-puromycin. Cells transfected with LV-NC lentivirus served as a negative control. Next, we used the hU6-MCS-CMV-puromycin plasmid to knock-down HOTAIRM1, DNMT1, and DNMT3B in cells (sh-HOTAIRM1, sh-DNMT1, and sh-DNMT3B) before transfection in MCF7 and T47D cells, with sh-NC cells used as a negative control. All cells were cultured in a humidified incubator with 5% CO2 and 95% air at 37 ℃ (Thermo Scientific, Waltham, MA, USA). Target sequences for RNA interference are listed in Table S2.
Immunohistochemistry and Western blot (WB) assays
Immunohistochemical experiments were performed using an immunohistochemistry kit (Solarbio, Beijing, China), according to the manufacturer’s instructions. Stained specimens were observed and images were captured under a Leica microscope (Leica, Germany). Total proteins were extracted using a total protein extraction kit (KeyGen Biotech, Nanjing, China), and concentrations were determined using the BCA-100 Protein Quantitative Analysis Kit. WB was performed according to standard protocols. WB transfer solution, WB electrophoresis solution, primary antibody diluent, and secondary antibody diluent were purchased from Beyotime Biotechnology (China). All antibodies used for WB are listed in Table S3.
Determination of cell viability and migration
Cell viability was determined using the cell counting kit 8 (CCK8) assay kit, according to the manufacturer’s instructions. Briefly, 1×103 breast cancer cells were first seeded into 96-well plates after cell transfection, then the CCK8 reagent (Dojindo, Japan) was added to each well followed by a 1 h incubation at 37 ℃ at 5 different time points (0, 1, 2, 3, and 4 days). Cell viability was measured by detecting absorbance at 450 nm and an optical density (OD) value was obtained. For the cell cloning assay, after cell transfection, breast cancer cells clone formation assay were performed through limiting dilution. Briefly, breast cancer cells were seeded into a 6-well plate and cultured for 14 days in DMEM supplemented with 10% FBS at 37 ℃ in 5% CO2 humidified air incubators. The cells were then stained with crystal violet and counted. For the cell migration assay, 5×104 breast cancer cells were seeded into a transwell chamber, with culture medium containing 20% serum DMEM used in the lower layer and serum-free DMEM used in the upper chamber. Cells were stained with crystal violet and counted after a 48-h incubation.
Statistical analysis
All bioinformatics analyses were performed using packages implemented in R software (unless stated otherwise). Statistical analysis of all experimental data, including Student’s t-test and Spearman correlations, were performed in GraphPad Prism 8, with P<0.05 considered statistically significant.
Results
Identification of methylation-driven genes in BRCA
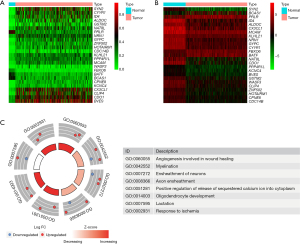
Profiles of methylation-driven genes in BRCA are shown using a flow diagram in Figure 1. A total of 5483 differentially expressed genes (DEGs) were identified based on FDR <0.05, log2FC >1, of which 3,216 and 2,267 were upregulated and downregulated in BRCA, respectively (available online: https://cdn.amegroups.cn/static/public/gs-22-164-01.xls). Since previous studies have demonstrated that overall promoter methylation levels are inversely correlated with gene expression (9-12), we next explored methylation-driven gene patterns by analyzing the DNA methylation data with DEG expression profiling, and identified a total of 25 methylation-driven genes based on FDR <0.05, log2FC >1, and cor <−0.3 (Table S4). Profiles of differential methylation levels and DEGs in the methylation-driven genes are shown in Figure 2A,2B. Next, we employed the “cluster profile”, “org.Hs.eg.db”, “enrichplot”, “ggplot2”, and “GOplot” packages in R to further investigate enriched pathways. Results from pathway analysis revealed significant enrichment of 8 Gene Ontology (GO) terms (GO:0060055, GO:0042552, GO:0007272, GO:0008366, GO:0051281, GO:0014003, GO:0007595, GO:0002931) (FDR <0.05) (Figure 2C) and 38 Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathways (Table S5). Collectively, these results suggested that gene expression levels were not only closely correlated with DNA methylation levels, but also play crucial roles in BRCA biology.
Construction of a prognostic model based on methylation-driven genes
Next, we analyzed the relationship between gene expression and the overall survival of breast cancer patients to understand the clinical relevance of these 25 methylation-driven genes. Firstly, we performed univariate Cox regression analysis on the training set and identified 6 prognosis-related genes (FDR <0.05) (Table S6). Next, we stratified patients into two groups, then subjected them to univariate and multivariate Cox proportional hazard regression analysis (Figure S1A). The resulting Kaplan-Meier survival curves suggested that the model had good performance, with individuals in the high-risk group associated with worse survival outcomes relative to those in the low-risk group (Figure 3A). Next, we performed univariate and multivariate Cox regression analyses to determine the relationship between clinicopathological features and the risk scores, and found that risk scores and each clinicopathological feature had good risk predictive ability (Figure S1B). However, results from multivariate analyses indicated that only the risk scores and age had excellent predictive ability for different clinical features (Figure 3B).
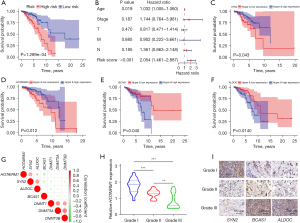
Further assessment of the association of methylation and expression with patient prognosis, based on Kaplan-Meier survival analysis, indicated that hypomethylation/high-expression genes (SYN2 and HOTAIRM1) (Figure 3C,3D) as well as hypermethylation/low-expression genes (BCAS1 and ALDOC) were significantly associated with patient prognosis (Figure 3E,3F). Moreover, DNA methylation transferases were negatively correlated with gene expression (Figure 3G). Validation of the 4 methylation-driven genes in clinical BRCA samples, based on qRT-PCR and immunohistochemical analyses, showed that the levels of SYN2 and HOTAIRM1 expression were negatively correlated with BRCA stage, whereas those of BCAS1 and ALDOC expression were positively correlated with BRCA stage (Figure 3H,3I). Collectively, these results indicated the successful construction of a methylation-driven gene risk prediction model for estimating the risk of BRCA.
Relationship between methylation-driven gene expression and DNA methylation levels
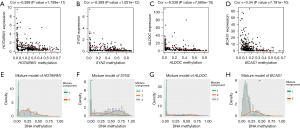
The correlation between the DNA methylation levels and expression of 25 methylation-driven genes in BRCA revealed a negative relationship between them (Figure 4A-4D, Figure S2). Moreover, results from the analysis of methylation levels of promoters of the 25 methylation-driven genes in BRCA and normal tissues indicated that most of the methylation-driven genes were hypermethylated in BRCA patients and hypomethylated in the normal group. On the other hand, FBXO6, BATF, BCAS1, and PRLR were hypomethylated in BRCA patients but hypermethylated in the normal group (Figure 4E-4H, Figures S3,S4). Collectively, these results suggested that BRCA patients were associated with high DNA methylation of methylation-driven genes.
Downregulation of HOTAIRM1 promotes the proliferation and migration of BRCA cells
We investigated the role of HOTAIRM1 in BRCA, owing to the fact that lncRNAs play an essential role in the proliferation and invasion of tumor cells. Firstly, we knocked down the expression of HOTAIRM1 in MCF7 and T47D cells, and verified the knockdown efficiency via qRT-PCR (Figure 5A). Next, we performed CCK8 and cell clonogenic found that downregulation of HOTAIRM1 promoted both the proliferation and clonal formation ability (Figure 5B-5D). Furthermore, we overexpressed HOTAIRM1 in MCF7 and T47D cells to determine its effect in BRCA, and verified the overexpression efficiency via qRT-PCR (Figure 5E). Results suggested that upregulation of HOTAIRM1 suppressed the proliferation and clonal formation ability of tumor cells (Figure 5F-5H). Parallelly, the invasion of MCF7 and T47D cells was promoted by down-regulation of HOTAIRM1, also inhibited by up-regulation of HOTAIRM1 expression (Figure 5I,5J). Finally, we performed a WB assay to determine the levels of proliferation-related (Cyclin E1 and Cyclin D1) and invasion-related (Vimentin, N-cadherin, E-cadherin) proteins (Figure 5K,5L). These findings suggested that HOTAIRM1 expression may be closely correlated with the proliferation and invasion of BRCA cells.
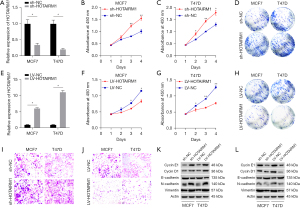
HOTAIRM1 expression is regulated by DNMT1 and DNMT3B
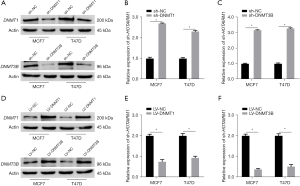
Next, we analyzed the correlation between methylation-driven gene expression and 3 methylation transferases (DNMT1, DNMT3A, and DNMT3B) and found that HOTAIRM1 was negatively correlated with methylation transferases DNMT1 and DNMT3B in BRCA (Figure 3G). To understand whether DNMT1 or DNMT3B also regulated HOTAIRM1 expression, we performed qRT-PCR and WB analyses in MCF7 and T47D cells transfection efficiency of silencing DNMT1 and DNMT3B (Figure 6A, Figure S5A-S5D). Results showed that HOTAIRM1 was significantly upregulated in cells that had DNMT1 or DNMT3B knocked down (Figure 6B,6C), We also performed qRT-PCR and WB analyses in MCF7 and T47D cells transfection efficiency of overexpressing DNMT1 and DNMT3B (Figure 6D), but downregulated in cells overexpressing DNMT1 and DNMT3B (Figure 6E,6F). Further evaluation of the association between HOTAIRM1 and DNMT1 or DNMT3B revealed a negative correlation in BRCA (Figure S5E,S5F). Collectively, these results demonstrated that DNMT1 and DNMT3B regulate the expression of HOTAIRM1.
Discussion
BRCA is the most common type of carcinoma among women (1,2,18). The current annual increase in BRCA incidence has necessitated the exploration of the specific molecular mechanisms underlying disease development and progression, as this will aid in the identification of effective prognostic biomarkers for predicting the survival rates of patients.
Previous studies have shown that DNA methylation plays a crucial role in regulating gene expression, while abnormal distribution of DNA methylation has been observed in many malignancies, including BRCA (13,14). Moreover, DNA methylation status has been strongly associated with the prognosis of BRCA patients (16,18). In the present study, we used a combination of methylation and transcriptome analyses to identify BRCA-specific diagnostic biomarkers for predicting survival rates.
We identified a total of 25 methylation-driven genes, then explored their clinical relevance by correlating their expression profiles with the overall survival of BRCA patients. Results from univariate and multivariate Cox regression analyses showed that hypomethylation/high-expression genes (SYN2 and HOTAIRM1) as well as hypermethylation/low-expression genes (BCAS1 and ALDOC) were significantly associated with patient prognosis. Additionally, results from distribution analysis of the degree of methylation indicated that most of the methylation-driven genes were hypermethylated in BRCA patients but hypomethylated in subjects in the normal group, while FBXO6, BATF, BCAS1, and PRLR were hypomethylated in BRCA patients but hypermethylated in subjects in the normal group. Among the identified methylation-driven genes, only HOTAIRM1 was a lncRNA. Much research evidence has described the roles played by lncRNAs in tumors, including cell proliferation, differentiation, chromosomal remodeling, epigenetic regulation, and transcription and post-transcriptional modification (22,23,25). For example, Kim et al. (29) demonstrated that HOTAIRM1 promotes tamoxifen resistance by mediating HOXA1 expression in ER+ breast cancer cells, indicating that it plays a vital role in the breast cancer cells. To date, however, no study has described the relationship between HOTAIRM1 and DNA methylation in BRCA.
For this reason, much attention has been directed towards HOTAIRM1’s role as a prognostic factor. In this research, we found that knockdown HOTAIRM1 expression promoted breast cancer cells proliferation, clone formation, and invasion. Up-regulation of HOTAIRM1 inhibited breast cancer cells proliferation, clone formation, and invasion. The expression of HOTAIRM1 is regulated by diverse and sophisticated mechanisms. Such as, inhibiting KDM6A demethylase represses HOTAIRM1 transcription, in addition IRF4 transcriptionally activate HOTAIRM1 (32,33). We also analyzed the relationship between methylation-driven gene expression and 3 methylation transferases (DNMT1, DNMT3A, and DNMT3B) and found that HOTAIRM1 was negatively correlated with DNMT1 and DNMT3B in BRCA. Overall, our results indicated that HOTAIRM1 was not only downregulated and hypermethylated in BRCA, but also plays a critical role in the proliferation and metastasis of breast cancer cells.
In conclusion, downregulation of HOTAIRM1 is a significant prognostic factor for the survival of BRCA patients, and thus may be a potential therapeutic target for the management of the disease.
Acknowledgments
We thank all of the participants in the present research. We also thank the reviewers for their valuable advice.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://gs.amegroups.com/article/view/10.21037/gs-22-164/rc
Data Sharing Statement: Available at https://gs.amegroups.com/article/view/10.21037/gs-22-164/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://gs.amegroups.com/article/view/10.21037/gs-22-164/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The present study was approved by the Ethics Committee of the First Affiliated Hospital of Gannan Medical University (No. LLSC-2022033101). Informed consent was obtained from patients or guardians.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Mannu GS, Wang Z, Broggio J, et al. Invasive breast cancer and breast cancer mortality after ductal carcinoma in situ in women attending for breast screening in England, 1988-2014: population based observational cohort study. BMJ 2020;369:m1570. [Crossref] [PubMed]
- Markham MJ, Wachter K, Agarwal N, et al. Clinical Cancer Advances 2020: Annual Report on Progress Against Cancer From the American Society of Clinical Oncology. J Clin Oncol 2020;38:1081. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Cuzick J, Sestak I, Forbes JF, et al. Use of anastrozole for breast cancer prevention (IBIS-II): long-term results of a randomised controlled trial. Lancet 2020;395:117-22. [Crossref] [PubMed]
- Padmanaban V, Krol I, Suhail Y, et al. E-cadherin is required for metastasis in multiple models of breast cancer. Nature 2019;573:439-44. [Crossref] [PubMed]
- Ahern TP, Broe A, Lash TL, et al. Phthalate Exposure and Breast Cancer Incidence: A Danish Nationwide Cohort Study. J Clin Oncol 2019;37:1800-9. [Crossref] [PubMed]
- Schmitz RJ, Lewis ZA, Goll MG. DNA Methylation: Shared and Divergent Features across Eukaryotes. Trends Genet 2019;35:818-27. [Crossref] [PubMed]
- Zhao S, Allis CD, Wang GG. The language of chromatin modification in human cancers. Nat Rev Cancer 2021;21:413-30. [Crossref] [PubMed]
- Zhou S, Zeng H, Huang J, et al. Epigenetic regulation of melanogenesis. Ageing Res Rev 2021;69:101349. [Crossref] [PubMed]
- Li Y, Chen X, Lu C. The interplay between DNA and histone methylation: molecular mechanisms and disease implications. EMBO Rep 2021;22:e51803. [Crossref] [PubMed]
- Martisova A, Holcakova J, Izadi N, et al. DNA Methylation in Solid Tumors: Functions and Methods of Detection. Int J Mol Sci 2021;22:4247. [Crossref] [PubMed]
- Do WL, Gohar J, McCullough LE, et al. Examining the association between adiposity and DNA methylation: A systematic review and meta-analysis. Obes Rev 2021;22:e13319. [Crossref] [PubMed]
- Parry A, Rulands S, Reik W. Active turnover of DNA methylation during cell fate decisions. Nat Rev Genet 2021;22:59-66. [Crossref] [PubMed]
- Bates SE. Epigenetic Therapies for Cancer. N Engl J Med 2020;383:650-63. [Crossref] [PubMed]
- Sun X, Yi J, Yang J, et al. An integrated epigenomic-transcriptomic landscape of lung cancer reveals novel methylation driver genes of diagnostic and therapeutic relevance. Theranostics 2021;11:5346-64. [Crossref] [PubMed]
- Győrffy B, Bottai G, Fleischer T, et al. Aberrant DNA methylation impacts gene expression and prognosis in breast cancer subtypes. Int J Cancer 2016;138:87-97. [Crossref] [PubMed]
- Xu Z, Sandler DP, Taylor JA. Blood DNA Methylation and Breast Cancer: A Prospective Case-Cohort Analysis in the Sister Study. J Natl Cancer Inst 2020;112:87-94. [Crossref] [PubMed]
- Boyne DJ, O'Sullivan DE, Olij BF, et al. Physical Activity, Global DNA Methylation, and Breast Cancer Risk: A Systematic Literature Review and Meta-analysis. Cancer Epidemiol Biomarkers Prev 2018;27:1320-31. [Crossref] [PubMed]
- Dietrich D, Lesche R, Tetzner R, et al. Analysis of DNA methylation of multiple genes in microdissected cells from formalin-fixed and paraffin-embedded tissues. J Histochem Cytochem 2009;57:477-89. [Crossref] [PubMed]
- Yballe CM, Vu TH, Hoffman AR. Imprinting and expression of insulin-like growth factor-II and H19 in normal breast tissue and breast tumor. J Clin Endocrinol Metab 1996;81:1607-12. [PubMed]
- Li S, Rong M, Iacopetta B. DNA hypermethylation in breast cancer and its association with clinicopathological features. Cancer Lett 2006;237:272-80. [Crossref] [PubMed]
- Statello L, Guo CJ, Chen LL, et al. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 2021;22:96-118. [Crossref] [PubMed]
- Rinn JL, Chang HY. Long Noncoding RNAs: Molecular Modalities to Organismal Functions. Annu Rev Biochem 2020;89:283-308. [Crossref] [PubMed]
- Gil N, Ulitsky I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat Rev Genet 2020;21:102-17. [Crossref] [PubMed]
- Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nat Cell Biol 2019;21:542-51. [Crossref] [PubMed]
- Sun H, Wang G, Peng Y, et al. H19 lncRNA mediates 17β-estradiol-induced cell proliferation in MCF-7 breast cancer cells. Oncol Rep 2015;33:3045-52. [Crossref] [PubMed]
- Wang J, Xie S, Yang J, et al. The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J Hematol Oncol 2019;12:81. [Crossref] [PubMed]
- Li D, Chai L, Yu X, et al. The HOTAIRM1/miR-107/TDG axis regulates papillary thyroid cancer cell proliferation and invasion. Cell Death Dis 2020;11:227. [Crossref] [PubMed]
- Kim CY, Oh JH, Lee JY, et al. The LncRNA HOTAIRM1 Promotes Tamoxifen Resistance by Mediating HOXA1 Expression in ER+ Breast Cancer Cells. J Cancer 2020;11:3416-23. [Crossref] [PubMed]
- Cedoz PL, Prunello M, Brennan K, et al. MethylMix 2.0: an R package for identifying DNA methylation genes. Bioinformatics 2018;34:3044-6. [Crossref] [PubMed]
- Gao C, Zhuang J, Li H, et al. Exploration of methylation-driven genes for monitoring and prognosis of patients with lung adenocarcinoma. Cancer Cell Int 2018;18:194. [Crossref] [PubMed]
- Li L, Deng J, Huang T, et al. IRF4 transcriptionally activate HOTAIRM1, which in turn regulates IRF4 expression, thereby affecting Th9 cell differentiation and involved in allergic rhinitis. Gene 2022;813:146118. [Crossref] [PubMed]
- Bah I, Youssef D, Yao ZQ, et al. Inhibiting KDM6A Demethylase Represses Long Non-Coding RNA Hotairm1 Transcription in MDSC During Sepsis. Front Immunol 2022;13:823660. [Crossref] [PubMed]
(English Language Editor: C. Betlazar-Maseh)


