
A comprehensive analysis of the diagnostic and prognostic value associated with the SLC7A family members in breast cancer
Introduction
Breast cancer (BC) is the most common type of cancer in women (1). It is a heterogeneous disease with major variations in incidence, clinical outcome, prognosis, and response to therapy (2). There are various subtypes of BC depending on the expression of the estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2), and histological grade (3). According to global cancer data from 2020, BC has replaced lung cancer as the most prevalent cancer worldwide, with 2.26 million cases compared to 2.2 million cases of lung cancer for the same year (4).
The Warburg effect is widely accepted as a common feature of metabolic reprogramming (5). However, accumulating evidence suggests that tumor cells depend on aerobic glycolysis as well as mitochondrial metabolism (6,7). Tumorigenesis depends on the reprogramming of cellular metabolism as both direct and indirect consequences of oncogenic mutations (8). The main nutrients consumed by mammalian cell proliferation are glucose and Amino acids and the conversion of glucose to lactate may help the amino acid divide into more new cells (9). For this reprogramming, tumor cells may regulate amino acid uptake by modulating the level or activity of specific amino acid transporters (10). Therefore, metabolic reprogramming based on these transporters may be a promising therapeutic target (11).
The SLC (Solute Carrier) series contains 65 SLC gene families with 458 different human transporter genes (12). SLC7A is an amino acid transporter/glycoprotein related family which is divided into two subgroups: the cationic amino acid transporters (the CAT family, SLC7A1–4 and SLC7A14) and the glycoprotein-associated amino acid transporters (the gpaAT family, SLC7A5–13) (13,14). Most members of the CAT subfamily have functional characteristics that promote diffuser function and mediate the entry and outflow of cationic amino acids (15). The gpaAT family, as known as an L-type amino acid transporter (LATs), is the heavy subunits and disulfide-linked heterodimers with the catalytic transporter subunits (16). The inhibition of solute carrier (SLC)7A5-mediated leucine influx has been associated with the downregulation of glycolytic metabolism, resulting in cancer-associated metabolic reprogramming, indicating that this molecule has a regulatory role in glycolysis reprogramming (17,18). SLC7A1 (Cat-1) regulates the survival of human breast cancer cell lines (19). In humans, moderate SLC7A3 expression has been observed in the brain, uterus, mammary gland, and testis (20). High SLC7A5 expression is associated with poor prognosis in different types of human cancers (21), including BC (22). However, the functional and prognostic significance of the other SLC7A family genes in BC remains unclear.
We hypothesized that the gene expression of the SLC7A family may be strongly associated with BC. Therefore, this study used multiple bioinformatics databases to analyze the expression of the SLC7A family genes in BC patients and evaluated their prognostic value. We present the following article in accordance with the REMARK reporting checklist (available at https://gs.amegroups.com/article/view/10.21037/gs-21-909/rc).
Methods
Ethics statement
All datasets were derived from public databases. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). As no patients were recruited in our study, ethics approval was not required.
Oncomine analysis
The SLC7A family transcript levels in different cancers were downloaded from the Oncomine website (https://www.oncomine.org/resource/login.html) and analyzed using Student’s t-test. A P value <0.01 and a fold change ≥2.0 were used as cutoff values.
Gene Expression Profiling Interactive Analysis (GEPIA) dataset
The correlation between the expression of the SLC7A family genes and the clinicopathological stages of patients and tumors were analyzed using the GEPIA (http://gepia.cancer-pku.cn/), web-based tool. Provisional datasets from TCGA and Genotype-Tissues Expression consortia were used.
The Cancer Genome Atlas (TCGA) and the Kaplan-Meier (KM) plotter database analyses
BC RNA-sequencing-based gene expression data (“Level_3_HTSeq-FPKM _normalized”) and patient survival data were downloaded from TCGA (https://portal.gdc.cancer.gov/). and used as the training dataset Data from the KM plotter database was used as the test dataset. The RNA-sequencing data were converted from fragments per kilobase of transcript per million mapped reads (FPKM) format to the transcripts per kilobase million mapped reads (TPM) format using log2 (TPM+1). A total of 1062 BC samples were categorized based on median RNA-sequencing data (TPM). Patients were grouped into a “high” or “low” signature expression group based on the median signature expression score. The ggplot2 package from the R software (version 3.6.3) was used to visualize the results. The KM plotter database was used for validation (23,24). Duplicate and redundant samples were removed and any biases between arrays were excluded.
Immunohistochemistry
Tissue processing and embedding was performed by the Department of Pathology, The Third Hospital of Nanchang. Antigen retrieval was performed by boiling the slides in a steamer with citrate buffer (pH 6.0, 95 °C) for 15 minutes. The 3-mm tumor sections were incubated with a rabbit anti-SLC7A5 antibody (1:500 dilution) and an anti-SLC7A4 antibody (1:200 dilution) at 4 °C overnight. Slides were then rinsed 3 times in phosphate buffered saline (PBS) and incubated with a secondary antibody at room temperature for 10 minutes, then counterstained with DAB for 2 minutes, followed by hematoxylin for 40 seconds. The above washing steps were repeated at each round of staining. Positively stained tissues were visualized by light microscopy (Olympus 600 auto-biochemical analyzer, Tokyo, Japan). As an additional control, the same procedure was performed without addition of primary antibodies.
Statistical analysis
Cox regression and survival analyses were performed using the R survival package (v 3.2-10), and the survminer package (version 0.4.9) was used for visualization. The rms package (6.2-0) was used to generate a nomogram for predicting overall survival (OS), progression-free interval (PFI), and disease-specific survival (DSS) in BC. A formal test of heterogeneity among subgroups was performed for each subgroup analysis. A P value <0.05 was considered statistically significant. Samples with missing clinical data were excluded from the multivariable analysis and nomograms. Prognostic data were obtained from a previous study (25).
Statistical analysis was performed in R (v.3.6.3), using pROC (version 1.17.0.1) for receiver operating characteristics (ROC) statistics and ggplot2 (v.3.3.3) for generating figures. A ROC curve was generated to evaluate the diagnostic performance of the SLC7A family.
Gene Set Enrichment Analysis (GSEA)
The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and Gene Ontology (GO) analysis for functional annotation of gene sets were performed using the R package cluster-Profiler (26). GO functional annotations in the categories biological process (BP), cellular component (CC), and molecular function (MF) were used to elucidate the biological implications of differential gene expression. GSEA (27) was performed using C2.CP.v.7.2 gene set collections from the Molecular Signature Database (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp) and cluster-Profiler (version 3.14.3) (26). Terms with false discovery rate (FDR) <0.25 and Padjust<0.05 were considered significant.
Analysis of the correlation between immune responses and the expression of the SLC7A family
ESTIMATE gives a measure of the immune-cell infiltrates using RNA-sequencing data through single-sample (ss) GSEA based on an inferred immune signature. The algorithm calculates the relative overexpression of immune cell types by implementing the “GSVA” function from the GSVA package (v.1.34.0) (28) in R (v.3.6.3) with method specification ssGSEA. Correlation analysis of immune response and the expression of the SLC7A family genes was performed using Spearman correlation. In total, 24 reported immune cell markers were evaluated (29).
Results
The levels of the SLC7A family transcripts in breast tumors
The expression of the SLC7A gene family was examined in various cancers by comparing normal tissues with tumor tissues using the Oncomine data (30-32) (Figure 1, Table 1). SLC7A1, SLC7A5, and SLC7A8 levels were significantly upregulated in human BC (P<0.05). SLC7A9, SLC7A13, and SLC7A14 were not expressed in normal breast tissues (P<0.05), whereas SLC7A3 and SLC7A4 expression levels were substantially decreased compared to normal tissues (P<0.001).
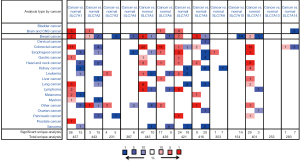
Table 1
| Gene | Type of breast cancer versus normal breast tissue | Fold change | P value | t-test | Source and/or reference |
|---|---|---|---|---|---|
| SLC7A1 | Medullary breast carcinoma | 2.24 | 1.15E-18 | 13.467 | (30) |
| Intraductal cribriform breast adenocarcinoma | 2.25 | 2.06E-5 | 9.58 | TCGA Breast | |
| SLC7A2 | Mucinous breast carcinoma | 3.017 | 6.82e-10 | 7.285 | (30) |
| SLC7A3 | NA | NA | NA | NA | NA |
| SLC7A4 | NA | NA | NA | NA | NA |
| SLC7A5 | Medullary breast carcinoma | 7.855 | 5.00E-16 | 13.504 | (30) |
| Invasive breast carcinoma | 3.301 | 2.20E-7 | 6.984 | (30) | |
| Invasive ductal breast carcinoma | 3.239 | 5.80E-67 | 24.518 | (30) | |
| Ductal breast carcinoma | 3.620 | 1.45E-6 | 6.000 | (31) | |
| Invasive breast carcinoma | 2.683 | 3.06E-15 | 8.797 | TCGA Breast | |
| Invasive ductal breast carcinoma | 3.443 | 1.65E-28 | 14.459 | TCGA Breast | |
| SLC7A6 | NA | NA | NA | NA | NA |
| SLC7A7 | Medullary breast carcinoma | 2.885 | 2.99E-10 | 8.521 | (30) |
| Invasive breast carcinoma stroma | 10.897 | 2.71E-12 | 14.008 | (32) | |
| SLC7A8 | Invasive breast carcinoma stroma | 4.259 | 1.07E-25 | 19.646 | (32) |
| Mixed lobular and ductal breast carcinoma | 3.448 | 1.55E-6 | 8.153 | TCGA Breast | |
| Mucinous breast carcinoma | 2.323 | 3.15E-14 | 10.145 | (30) | |
| SLC7A9 | NA | NA | NA | NA | NA |
| SLC7A10 | NA | NA | NA | NA | NA |
| SLC7A11 | Ductal breast carcinoma | 2.940 | 4.28E-5 | 5.063 | (31) |
| Invasive ductal breast carcinoma | 2.943 | 8.06E-18 | 10.233 | TCGA Breast | |
| SLC7A13 | NA | NA | NA | NA | NA |
| SLC7A14 | NA | NA | NA | NA | NA |
NA, not available; TCGA, The Cancer Genome Atlas.
The correlation between the SLC7A family mRNA levels and the clinicopathological stages of BC patients
The mRNA levels of the SLC7A family in BC and normal breast tissues was compared using the TCGA data. SLC7A5, SLC7A7, SLC7A8, SLC7A11, and SLC7A13 mRNA levels were substantially higher in BC compared to normal breast tissue (P<0.05). In addition, SLC7A1 and SLC7A9 mRNA levels were significantly higher in BC tissue compared to normal breast tissue (P<0.01). However, SLC7A3, SLC7A4, SLC7A10, and SLC7A14 mRNA levels were lower in BC tissues compared to normal tissues (P<0.01) (Figure 2).

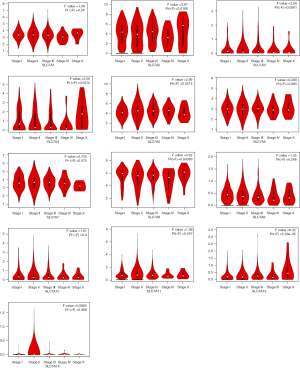
Using the GEPIA dataset, the correlation between the expression of the SLC7A family members and BC tumor staging was analyzed. SLC7A2, SLC7A4, SLC7A5, and SLC7A13 were significantly positively correlated with tumor stages, whereas the other family members did not significantly differ in expression among tumor stages (Figure 3).
The correlation between SLC7A family mRNA levels and improved prognosis of BC patients
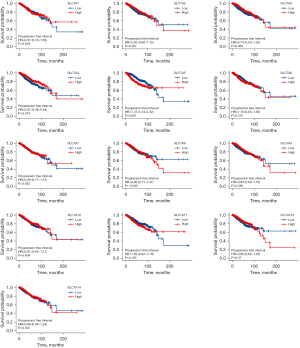
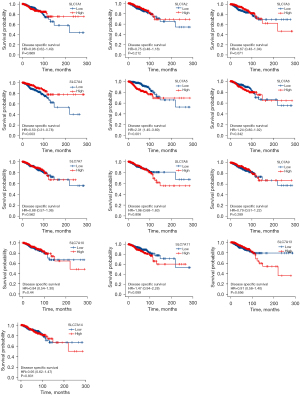
To date, only a few members of the SLC7A family have been investigated for potential prognostic value in the TCGA cohort. As SLC7A12 has only been discovered in mice to date, it was not included in the bioinformatics analysis. KM analysis of prognosis was performed for the SLC7A family members using the TCGA data. The results of the Cox regression modeling showed that high SLC7A3 expression could be considered a significant prognostic marker for OS time in BC patients, with a hazard ratio (HR) of 0.59 and log rank P of 0.001. The median survival time of patients with high SLC7A3 expression was 131.5 months (Figure 4). High SLC7A5 expression in BC patients indicated a poorer survival time (HR =1.72, log rank P=0.001), and the median follow-up for patients was 116.4 months. The other SLC7A family members were not correlated with OS (Figure 5). For PFI, SLC7A4 (HR =0.67, P=0.019) and SLC7A5 (HR =1.73, P=0.001) were identified as prognostic biomarkers (Figure 5). For DSS, SLC7A4 (HR =0.50, P=0.003) and SLC7A5 (HR =2.31, P<0.001) were identified as prognostic biomarkers (Figure 6).
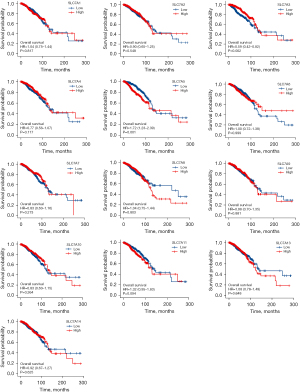
In the KM plotter database, analysis indicated that higher than median expression levels of SLC7A4 (HR =0.83, log rank P=0.048) and SLC7A5 (HR =1.64, log rank P=2.4e-07) were correlated with favorable OS for BC. For SLC7A3, the result was not significant (HR =0.78, log rank P=0.65) (Figure 7A-7C).
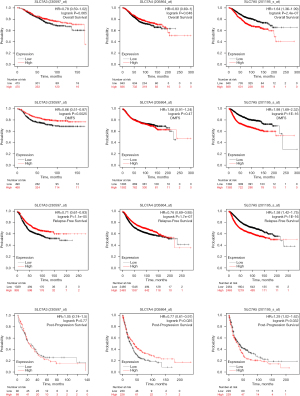
SLC7A5 (HR =1.98, log rank P<1e-16) indicated a poorer distant metastasis-free survival (DMFS) time in patients with BC. High expressions of SLC7A3 revealed a good DMFS (HR =0.66, log rank P=0.0025) (Figure 7D-7F).
High expressions of SLC7A3 (HR =0.71, log rank P=1.1e-05) and SLC7A4 (HR =0.76, log rank P=1.7e-07), indicated good relapse-free survival (RFS) (Figure 7G-7I).
High SLC7A5 expression indicated a poor post-progression survival (PPS) time (HR =1.29, log rank P=0.032), whereas high SLC7A4 expression indicated a good PPS time in patients with BC (HR =0.69, log rank P=0.041, HR =0.77, log rank P=0.025, respectively) (Figure 7J-7L).
Interestingly, examination of the TCGA and KM plotter datasets revealed that high expression of SLC7A4 and SLC7A5, and possibly SLC7A3, in breast tumors is associated with survival progression.
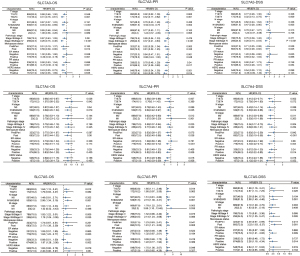
Thus, exploratory subgroup analyses were performed for these 3 genes in terms of OS, PFI, and DSS, as well as for TNM factors, menopausal status, ER status, PR status, HER2 status, and tumor stage. Prespecified subgroups were compared via Cox regression analysis (Table 2). KM analyses showed that high expression of SLC7A3 and SLC7A4 was associated with better OS compared to low expression of these genes in certain BC subgroups, including T3, 4, N1, 2, 3, and M0; stages III and IV; ER-positive; PR-positive; and HER2-negative patients (Figure 8). Moreover, in patients with high SLC7A5 expression, the T1, 2, N1, 2, 3, and M0 subgroups, the stages I and II subgroups; the ER-positive subgroup, and the PR-positive subgroup all showed very poor prognosis. However, there were no statistically significant associations in the HER2 status subgroup (Figure 8).
Table 2
| Characteristics | Total (N) | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | |||
| T stage (T3 & T4 vs. T1 & T2) | 1,061 | 1.673 (1.152–2.429) | 0.007 | 1.288 (0.639–2.596) | 0.478 | |
| N stage (N1 & N2 & N3 vs. N0) | 1,045 | 2.145 (1.497–3.073) | <0.001 | 1.009 (0.505–2.019) | 0.979 | |
| Pathologic stage (Stage III & Stage IV vs. Stage I & Stage II) | 1,041 | 2.519 (1.787–3.549) | <0.001 | 2.906 (1.353–6.240) | 0.006 | |
| Menopause status (post vs. pre & peri) | 955 | 2.405 (1.445–4.002) | <0.001 | 4.301 (2.032–9.101) | <0.001 | |
| ER status (positive vs. negative) | 1,014 | 0.704 (0.487–1.017) | 0.062 | 0.503 (0.287–0.884) | 0.017 | |
| PR status (positive vs. negative) | 1,011 | 0.762 (0.541–1.074) | 0.120 | |||
| HER2 status (positive vs. negative) | 705 | 1.611 (0.981–2.644) | 0.059 | 1.142 (0.611–2.133) | 0.677 | |
ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2.
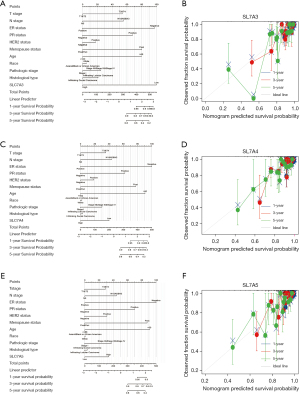
A nomogram integrating SLC7A3, SLC7A4, and SLC7A5 expression and independent clinical risk factors (TN pathological stage, ER, PR, HER2, and menstrual status) was constructed based on 1000 bootstrap replicates (Figure 9). The higher the total number of points on the nomogram, the worse the prognosis. The C-index values were 0.820 for SLC7A3, 0.784 for SLC7A4, and 0.792 for SLC7A5. The deviation correction line in the calibration plot was close to the ideal curve (i.e., a 45° line), indicating that the prediction model outputs were in good agreement with the observed results (Figure 9).
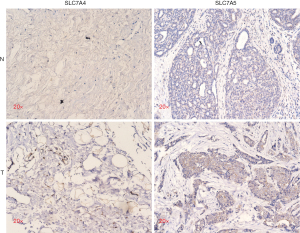
Furthermore, an immunohistochemical (IHC) assay was developed to examine SLC7A4 and SLC7A5 expression in formalin-fixed BC tissue. SLC7A5 expression in tumor tissues was found to be significantly higher than that in normal breast tissues. Conversely, SLC7A4 expression in normal tissues was significantly higher than that in breast tumor tissues (Figure 10).
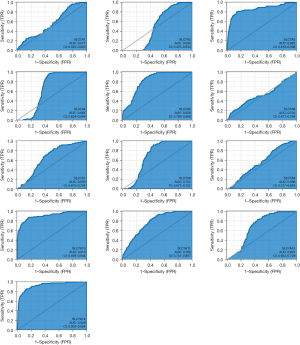
Diagnostic value of the SLC7A family genes in BC patients
The predictive value of each SLC7A family member was evaluated in BC patients based on the AUC using the TCGA data. SLC7A10 and SLC7A14 had outstanding diagnostic value in distinguishing BC patients from healthy individuals (AUC 0.910 and 0.926, respectively). SLC7A3, SLC7A5, SLC7A8, and SLC7A11 also showed excellent diagnostic value (with AUC values of 0.862, 0.828, 0.702, and 0.769, respectively). SLC7A1, SLC7A2, SLC7A4, SLC7A6, SLC7A7, SLC7A9, and SLC7A13 had moderate prognostic value for distinguishing BC patients (AUC values of 0.571, 0.509, 0.664, 0.532, 0.659, 0.566, and 0.691, respectively) (Figure 11).
Functional enrichment analysis of SLC7A family genes
GO enrichment analysis of the SLC7A genes that were significantly differentially expressed in BC revealed various over-represented terms in three functional groups, namely, CC, BPs, and MF (Figure 12A-12D).
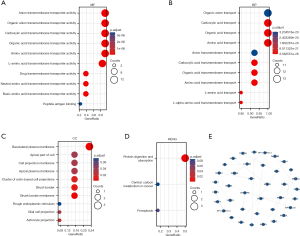
In the CC category, the SLC7A family genes that were upregulated in breast tumor tissues were primarily involved in basolateral plasma membrane, brush border membrane, brush border, cluster of actin-based cell projections, astrocyte projection, glial cell projection, apical plasma membrane, cell projection membrane, apical part of the cell, and rough endoplasmic reticulum. In the BP category, they were primarily involved in amino acid transport, amino acid transmembrane transport, L-alpha-amino acid transmembrane transport, L-amino acid transport, organic acid transmembrane transport, carboxylic acid transmembrane transport, organic acid transport, carboxylic acid transport, and anion transmembrane transport. According to KEGG pathways analysis, the genes were involved in protein digestion and absorption (SLC7A7, SLC7A9, and SLC7A8), ferroptosis (SLC7A11), and central carbon metabolism in cancer (SLC7A5). Figure 12E shows an interactive network of the most highly enriched terms (colored by cluster-ID, where distinct colors indicate enriched pathways).
SLC7A1 and SLC7A3-5-related signaling pathways were obtained via GSEA (Figure 13).
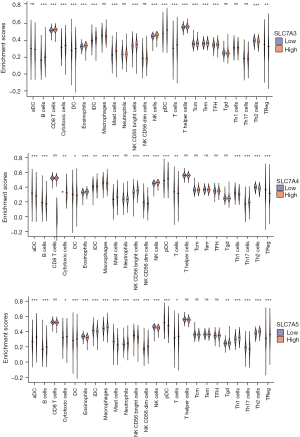
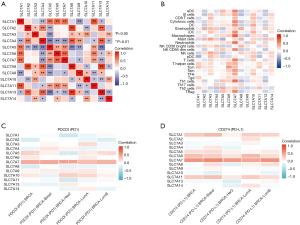
The correlation between SLC7A family gene expression and immune infiltration
Although the complex interactions between solid tumors and their microenvironment remain unclear, studies have shown that immune infiltrates are significantly related to the progression and prognosis of BC (33-35). A ssGSEA was conducted to determine the relative abundance of various immune cell types based on expression data from TCGA. Significant differences in relative SLC7A family gene expression were observed between tumor and normal tissues for immune cells (Figure 14). We found significant correlations among SLC7A family expression levels (Figure 15A). In addition, the correlation coefficients between SLC7A family member expression and immune-cell subsets were determined (Figure 15B). The results showed that SLC7A expression levels were closely correlated to 24 immune cell markers. T helper 2 (Th2) cells, natural killer (NK) CD56bright cells, and NK cells were the most strongly correlated with SLC7A family genes, suggesting that SLC7A family members play a role in BC by modulating immune cell infiltration. Notably, SLC7A7 expression was strongly positively correlated with immune-cell infiltration. Programmed cell death protein 1 (PDCD1 or PD-1) and programmed death-ligand 1 (PD-L1 or CD274) levels were positively correlated with SLC7A5, SLC7A6, and SLC7A7 expression (Figure 15C,15D).
Discussion
To the best of our knowledge, this study represents the first comprehensive evaluation of various large, well-established, and validated databases, including TCGA, and the KM database. We systematically documented the expression, prognostic value, relationship with immune infiltration and functional enrichment of the SLC7A family genes in BC. ROC analysis revealed that SLC7A family genes have a significant diagnostic value for BC and may play an important role in BC pathogenesis. Univariate survival analysis demonstrated that BC patients with high SLC7A5 expression had poor prognosis. In contrast, high SLC7A3 and SLC7A4 expression were associated with good prognosis. This data also provided a genomic explanation for the adverse prognosis associated with specific clinical, pathologic, and molecular features. It also confirmed that the occurrence of BC is highly dependent on glucose metabolism (7). Accumulating evidence suggests that SLC7A members that encode amino acid and glucose transporters may be therapeutic targets in cancers.
SLC7A1 has been suggested as a promising therapeutic target for colorectal cancers (36), hepatoblastomas (37), BC cell lines (19), pediatric sarcomas, and brain tumors (38).
SLC7A2 promotes the proliferation, migration, and invasiveness of ovarian cancer cells and was thus identified as a prognostic biomarker and potential therapeutic target for ovarian cancer (39,40). Differential gene expression analysis of different patient subgroups stratified based on apoptotic caspase expression suggested that SLC7A2 may be a prognostic biomarker with independent prognostic value in BC (41-43). In our survival analysis of the KM plotter database, significant differences were observed between expression levels of SLC7A2 and prognosis.
High expression of SLC7A3, SLC7A5, and SLC7A11 has been associated with poor prognosis in papillary thyroid carcinoma patients (44). In addition, SLC27A4 may be a new potential therapeutic target for lung tumor chemotherapy (45). Similarly, in our study, BC patients with high SLC7A4 expression had better prognosis. SLC7A5 expression is upregulated with tumor progression and correlates with poor clinical outcomes, and thus, has emerged as an important factor in cancer-associated metabolic reprogramming (46-48). In HER2-negative BC, LAT1 (SLC7A5) may serve as an important prognostic and predictive marker and a potential therapeutic target (49), as validated by our results. A growing body of evidence has demonstrated that metabolic reprogramming contributes to the acquisition and maintenance of robustness associated with malignancy (50,51). These findings are in line with our results. SLC7A5 plays a role in subtype-specific activation of immune-cell infiltration (52). mTOR signaling regulated by SLC7A5/SLC7A6 is a sensor of dynamic alterations in nutrients in the tumor microenvironment (53,54).
SLC7A7 plays a critical role in the pathogenesis of T-cell acute lymphoblastic leukemia (55). SLC7A7 variants have been associated with the risk of glioma in a Chinese population (56,57). SLC7A8 is a prognostic biomarker in ER-positive low-invasive BC (58).
Ferroptosis is an iron-dependent form of non-apoptotic cell death. Upregulation of SLC7A11 expression is highly dependent on glucose and glutamine levels, which may be therapeutic targets, and are associated with ferroptosis (59,60). In this current study, KEGG pathway analysis revealed that SLC7A11 is associated with ferroptosis.
Immunotherapy has been recognized as a promising cancer treatment, and T-cell tumor infiltration has been associated with effective immunotherapy in cancer patients (61). High expression of T-cell and B-cell signatures predicts improved OS in various tumor types, including BC (CD8 T cells, HR =0.36, 95% confidence interval: 0.16–0.81, P=0.01) (62). NK cells are an important mediator of the anti-tumor immune response and kill malignant cells (63). The current study demonstrated a correlation between SLC7A family gene expression and infiltrating immune cells. Thus, SLC7A5 and SLC7A7 may provide a rationale for immunotherapy in BC patients. The KEYNOTE-012 randomized phase Ib study of pembrolizumab in patients with advanced triple-negative BC suggested that the overall response rate was 18.5%, and the median time to response was 17.9 weeks (range, 7.3–32.4 weeks) (64). Our results suggested that SLC7A7 may be a candidate predictive biomarker for immunotherapy efficacy and is associated with prognosis in BC patients. SLC7A family genes in BC play an important role in the growth and survival of cancer, making potential new therapeutic targets. The application prospect is potential to provide new strategies for drug discovery, pharmacokinetics, drug delivery, drug elimination and medically related transporters in the future.
Although our findings improve the understanding of the relationship between the SLC7A family members and BC, there were some limitations to this study. First, the data were obtained from public databases, and although the data quality was uniform, it may not be completely rigorous. The number of male patients with tumor stage M were too small for subgroup analysis. Further experimental studies are needed to confirm our results. Second, the specific role of SLC7A in BC development should be comprehensively assessed, with consideration given to broader clinical factors. The current analyses were limited by the lack of certain information in public databases. Third, the correlation between SLC7A expression at the mRNA and protein levels should be verified in cellular experiments and clinical trials. Fourth, the precise mechanisms by which SLC7A contributes to BC development warrants further research.
In conclusion, this study provided some insights into the role of SLC7A in BC and identified potential diagnostic and prognostic biomarkers. Indeed, SLC7A5 expression may be a biomarker of poor prognosis in BC patients, while high SLC7A4 expression may suggest good prognosis. Overall, the SLC7A family has good diagnostic efficacy in BC. Finally, the SLC7A family members may be predictive biomarkers for determining the efficacy of immunotherapy and the prognosis of BC patients.
Acknowledgments
We would like to acknowledge the support provided by www.xiantao.love.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://gs.amegroups.com/article/view/10.21037/gs-21-909/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://gs.amegroups.com/article/view/10.21037/gs-21-909/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bundred NJ. Prognostic and predictive factors in breast cancer. Cancer Treat Rev 2001;27:137-42. [Crossref] [PubMed]
- Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med 2009;360:790-800. [Crossref] [PubMed]
- Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol 2011;5:5-23. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Warburg O, Wind F, Negelein E. THE METABOLISM OF TUMORS IN THE BODY. J Gen Physiol 1927;8:519-30. [Crossref] [PubMed]
- Bravatà V, Stefano A, Cammarata FP, et al. Genotyping analysis and 18FDG uptake in breast cancer patients: a preliminary research. J Exp Clin Cancer Res 2013;32:23. [Crossref] [PubMed]
- El Ansari R, McIntyre A, Craze ML, et al. Altered glutamine metabolism in breast cancer; subtype dependencies and alternative adaptations. Histopathology 2018;72:183-90. [Crossref] [PubMed]
- Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab 2016;23:27-47. [Crossref] [PubMed]
- Kim J, Kang J, Kang YL, et al. Ketohexokinase-A acts as a nuclear protein kinase that mediates fructose-induced metastasis in breast cancer. Nat Commun 2020;11:5436. [Crossref] [PubMed]
- Bhutia YD, Babu E, Ramachandran S, et al. Amino Acid transporters in cancer and their relevance to "glutamine addiction": novel targets for the design of a new class of anticancer drugs. Cancer Res 2015;75:1782-8. [Crossref] [PubMed]
- Yoshida GJ. Metabolic reprogramming: the emerging concept and associated therapeutic strategies. J Exp Clin Cancer Res 2015;34:111. [Crossref] [PubMed]
- Schaller L, Lauschke VM. The genetic landscape of the human solute carrier (SLC) transporter superfamily. Hum Genet 2019;138:1359-77. [Crossref] [PubMed]
- Fotiadis D, Kanai Y, Palacín M. The SLC3 and SLC7 families of amino acid transporters. Mol Aspects Med 2013;34:139-58. [Crossref] [PubMed]
- Verrey F, Closs EI, Wagner CA, et al. CATs and HATs: the SLC7 family of amino acid transporters. Pflugers Arch 2004;447:532-42. [Crossref] [PubMed]
- Closs EI, Boissel JP, Habermeier A, et al. Structure and function of cationic amino acid transporters (CATs). J Membr Biol 2006;213:67-77. [Crossref] [PubMed]
- Errasti-Murugarren E, Fort J, Bartoccioni P, et al. L amino acid transporter structure and molecular bases for the asymmetry of substrate interaction. Nat Commun 2019;10:1807. [Crossref] [PubMed]
- Yoon BR, Oh YJ, Kang SW, et al. Role of SLC7A5 in Metabolic Reprogramming of Human Monocyte/Macrophage Immune Responses. Front Immunol 2018;9:53. [Crossref] [PubMed]
- Kandasamy P, Gyimesi G, Kanai Y, et al. Amino acid transporters revisited: New views in health and disease. Trends Biochem Sci 2018;43:752-89. [Crossref] [PubMed]
- Abdelmagid SA, Rickard JA, McDonald WJ, et al. CAT-1-mediated arginine uptake and regulation of nitric oxide synthases for the survival of human breast cancer cell lines. J Cell Biochem 2011;112:1084-92. [Crossref] [PubMed]
- Vékony N, Wolf S, Boissel JP, et al. Human cationic amino acid transporter hCAT-3 is preferentially expressed in peripheral tissues. Biochemistry 2001;40:12387-94. [Crossref] [PubMed]
- Zhang C, Xu J, Xue S, et al. Prognostic Value of L-Type Amino Acid Transporter 1 (LAT1) in Various Cancers: A Meta-Analysis. Mol Diagn Ther 2020;24:523-36. [Crossref] [PubMed]
- Li Y, Wang W, Wu X, et al. SLC7A5 serves as a prognostic factor of breast cancer and promotes cell proliferation through activating AKT/mTORC1 signaling pathway. Ann Transl Med 2021;9:892. [Crossref] [PubMed]
- Györffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010;123:725-31. [Crossref] [PubMed]
- Győrffy B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput Struct Biotechnol J 2021;19:4101-9. [Crossref] [PubMed]
- Liu J, Lichtenberg T, Hoadley KA, et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018;173:400-416.e11. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [Crossref] [PubMed]
- Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013;39:782-95. [Crossref] [PubMed]
- Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012;486:346-52. [Crossref] [PubMed]
- Richardson AL, Wang ZC, De Nicolo A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell 2006;9:121-32. [Crossref] [PubMed]
- Finak G, Bertos N, Pepin F, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med 2008;14:518-27. [Crossref] [PubMed]
- Baxevanis CN, Sofopoulos M, Fortis SP, et al. The role of immune infiltrates as prognostic biomarkers in patients with breast cancer. Cancer Immunol Immunother 2019;68:1671-80. [Crossref] [PubMed]
- de Melo Gagliato D, Cortes J, Curigliano G, et al. Tumor-infiltrating lymphocytes in Breast Cancer and implications for clinical practice. Biochim Biophys Acta Rev Cancer 2017;1868:527-37. [Crossref] [PubMed]
- Zhu T, Zheng J, Hu S, et al. Construction and validation of an immunity-related prognostic signature for breast cancer. Aging (Albany NY) 2020;12:21597-612. [Crossref] [PubMed]
- Okita K, Hara Y, Okura H, et al. Antitumor effects of novel mAbs against cationic amino acid transporter 1 (CAT1) on human CRC with amplified CAT1 gene. Cancer Sci 2021;112:563-74. [Crossref] [PubMed]
- He W, Zhang J, Liu B, et al. S119N Mutation of the E3 Ubiquitin Ligase SPOP Suppresses SLC7A1 Degradation to Regulate Hepatoblastoma Progression. Mol Ther Oncolytics 2020;19:149-62. [Crossref] [PubMed]
- Vardon A, Dandapani M, Cheng D, et al. Arginine auxotrophic gene signature in paediatric sarcomas and brain tumours provides a viable target for arginine depletion therapies. Oncotarget 2017;8:63506-17. [Crossref] [PubMed]
- Sun T, Bi F, Liu Z, et al. SLC7A2 serves as a potential biomarker and therapeutic target for ovarian cancer. Aging (Albany NY) 2020;12:13281-96. [Crossref] [PubMed]
- Coburn LA, Singh K, Asim M, et al. Loss of solute carrier family 7 member 2 exacerbates inflammation-associated colon tumorigenesis. Oncogene 2019;38:1067-79. [Crossref] [PubMed]
- Rodriguez-Ruiz ME, Buqué A, Hensler M, et al. Apoptotic caspases inhibit abscopal responses to radiation and identify a new prognostic biomarker for breast cancer patients. Oncoimmunology 2019;8:e1655964. [Crossref] [PubMed]
- Kesavardhana S, Kanneganti TD. Targeting Apoptosis Inhibition to Activate Antitumor Immunity. Trends Immunol 2019;40:1073-5. [Crossref] [PubMed]
- Böckers M, Paul NW, Efferth T. Organophosphate ester tri-o-cresyl phosphate interacts with estrogen receptor α in MCF-7 breast cancer cells promoting cancer growth. Toxicol Appl Pharmacol 2020;395:114977. [Crossref] [PubMed]
- Shen L, Qian C, Cao H, et al. Upregulation of the solute carrier family 7 genes is indicative of poor prognosis in papillary thyroid carcinoma. World J Surg Oncol 2018;16:235. [Crossref] [PubMed]
- Wu S, Su J, Qian H, et al. SLC27A4 regulate ATG4B activity and control reactions to chemotherapeutics-induced autophagy in human lung cancer cells. Tumour Biol 2016;37:6943-52. [Crossref] [PubMed]
- Häfliger P, Charles RP. The L-Type Amino Acid Transporter LAT1-An Emerging Target in Cancer. Int J Mol Sci 2019;20:2428. [Crossref] [PubMed]
- Lu X. The Role of Large Neutral Amino Acid Transporter (LAT1) in Cancer. Curr Cancer Drug Targets 2019;19:863-76. [Crossref] [PubMed]
- Salisbury TB, Arthur S. The Regulation and Function of the L-Type Amino Acid Transporter 1 (LAT1) in Cancer. Int J Mol Sci 2018;19:2373. [Crossref] [PubMed]
- Bodoor K, Almomani R, Alqudah M, et al. LAT1 (SLC7A5) Overexpression in Negative Her2 Group of Breast Cancer: A Potential Therapy Target. Asian Pac J Cancer Prev 2020;21:1453-8. [Crossref] [PubMed]
- Yoshida GJ. The Harmonious Interplay of Amino Acid and Monocarboxylate Transporters Induces the Robustness of Cancer Cells. Metabolites 2021;11:27. [Crossref] [PubMed]
- El-Ansari R, Craze ML, Alfarsi L, et al. The combined expression of solute carriers is associated with a poor prognosis in highly proliferative ER+ breast cancer. Breast Cancer Res Treat 2019;175:27-38. [Crossref] [PubMed]
- Ansari RE, Craze ML, Althobiti M, et al. Enhanced glutamine uptake influences composition of immune cell infiltrates in breast cancer. Br J Cancer 2020;122:94-101. [Crossref] [PubMed]
- Wang Q, Holst J. L-type amino acid transport and cancer: targeting the mTORC1 pathway to inhibit neoplasia. Am J Cancer Res 2015;5:1281-94. [PubMed]
- Liu P, Ge M, Hu J, et al. A functional mammalian target of rapamycin complex 1 signaling is indispensable for c-Myc-driven hepatocarcinogenesis. Hepatology 2017;66:167-81. [Crossref] [PubMed]
- Ji X, Yang X, Wang N, et al. Function of SLC7A7 in T-Cell Acute Lymphoblastic Leukemia. Cell Physiol Biochem 2018;48:731-40. [Crossref] [PubMed]
- Fan S, Zhao Y, Li X, et al. Genetic variants in SLC7A7 are associated with risk of glioma in a Chinese population. Exp Biol Med (Maywood) 2013;238:1075-81. [Crossref] [PubMed]
- Fan S, Meng D, Xu T, et al. Overexpression of SLC7A7 predicts poor progression-free and overall survival in patients with glioblastoma. Med Oncol 2013;30:384. [Crossref] [PubMed]
- El Ansari R, Alfarsi L, Craze ML, et al. The solute carrier SLC7A8 is a marker of favourable prognosis in ER-positive low proliferative invasive breast cancer. Breast Cancer Res Treat 2020;181:1-12. [Crossref] [PubMed]
- Koppula P, Zhuang L, Gan B. Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy. Protein Cell 2021;12:599-620. [Crossref] [PubMed]
- Lin W, Wang C, Liu G, et al. SLC7A11/xCT in cancer: biological functions and therapeutic implications. Am J Cancer Res 2020;10:3106-26. [PubMed]
- Borst J, Ahrends T, Bąbała N, et al. CD4+ T cell help in cancer immunology and immunotherapy. Nat Rev Immunol 2018;18:635-47. [Crossref] [PubMed]
- Iglesia MD, Parker JS, Hoadley KA, et al. Genomic Analysis of Immune Cell Infiltrates Across 11 Tumor Types. J Natl Cancer Inst 2016;108:djw144. [Crossref] [PubMed]
- Wu SY, Fu T, Jiang YZ, et al. Natural killer cells in cancer biology and therapy. Mol Cancer 2020;19:120. [Crossref] [PubMed]
- Nanda R, Chow LQ, Dees EC, et al. Pembrolizumab in Patients With Advanced Triple-Negative Breast Cancer: Phase Ib KEYNOTE-012 Study. J Clin Oncol 2016;34:2460-7. [Crossref] [PubMed]
(English Language Editor: J. Teoh)



