
Upregulated circRAD18 promotes tumor progression by reprogramming glucose metabolism in papillary thyroid cancer
Introduction
Thyroid cancer is the fifth most common malignancy according to global tumor statistics released in 2020 (1). Papillary thyroid cancer is the most common pathological subtype, accounting for about 85% thyroid cancer patients (2), and with progress made in surgery, over 90% patients of patients can be completely cured after standard and systematic treatment (3,4). However, nearly 10% of thyroid cancers have distant metastases or local recurrences (5) and the overall survival of patients with recurrent and metastatic disease is the biggest obstacle to treatment (6). The aim of this study was to explore the underlying specific mechanism of papillary thyroid cancer growth and invasion.
Noncoding RNAs are vital regulators of cancer biology, among which circRNAs are a newly verified type of intracellular noncoding RNA (7). Having no 3’-tail or 5’-head structure, which are very abundant in many human cells, circ RNAs (8) have hundreds of thousands bases and are able to modulate the function or change the expression of many important genes by different mechanisms. These include binding with proteins, blocking microRNAs (miRNAs), and even encoding low molecular weight proteins (9). New technology in circRNA sequencing, including high-throughput microarray sequencing, has seen increasing numbers of circRNAs identified and proven as modulators of various disorders, including different kinds of cancers (10-14). Taking the most well-known circRNA as an example, CDR1AS/ciRS-7 facilitates the tumor proliferation, chemo-resistance, metastasis, and immune evasion of different cancers by blocking miRNA miR-7 (15-20). Circular RNA FBXW7 is one of the first genes discovered to encode proteins which can suppress cell proliferation and invasion by encoding a 21 kDa/185aa novel short peptide in glioma and breast cancer (21,22). Additionally, circular RNA RAD18 was discovered as a blocker of miR-208a in breast cancer (23). CircHIPK3 was proven to promote colorectal cancer progression by sponging miR-7 (24). However, the underlying specific mechanism of circRNAs in papillary thyroid cancer growth and invasion remains unknown.
Our results firstly showed circRAD18 was differentially overexpressed in papillary thyroid cancer tissues and cells, and interestingly, was also involved in glucose metabolism. In addition, molecular experiments uncovered the mechanism of the circRAD18-miR-516b-PDK1 axis in papillary thyroid cancer metabolic reprogramming progression.
We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/gs-21-481).
Methods
Clinical tissue collection
Fresh papillary thyroid cancer and adjacent thyroid tissues were collected from Sun Yat-sen University Cancer Center (SYSUCC). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of the SYSUCC (GZR2017163). Informed consent was obtained from all involved patients.
Cell line and culture
Cell lines were obtained from the ATCC (TPC-1, NPA87, and KAT-5) and were cultured in the medium (Gibco) containing 10% FBS (Gibco) according to the supplier’s instructions.
RT-qPCR analysis
TRIzol reagent (Invitrogen, USA) was used to extract RNA. The primers for circRAD18 were F: 5′-CAACAGCTCATTAAAAGGCA-3′, R: 5′-TACAGAGAGAGCAGTCTGAT-3′, for RAD18 were F: 5′-TCCTGTAGCCTCCAGACAGTC-3′, R: 5′-GCTCGCCTCTTTTTGAGGG-3′, and for GAPDH were F: 5′-CTGGGCTACACTGAGCACC-3′, R: 5′-AAGTGGTCGTTGAGGGCAATG-3′.
RNase R resistant assay
After 5 ug RNA was treated with RNase R (2 U/μg) or mock for half an hour at 37 °C, the remaining circRNA and other RNAs were quantified by RT-qPCR analysis.
Actinomycin D digestion experiment
Papillary thyroid cancer cells were treated with 5 μg/mL actinomycin D for certain hour period. Cells were then collected and the circular form of circRAD18 and linear form of RAD18 mRNA were quantified.
Western blot
Protein was extracted by RIPA lysis then loaded, separated, and transferred to PVDF membranes. The membrane was incubated with primary antibody anti-PDK1 (1:1,000, Cell Signaling, USA), and anti-GAPDH antibody (1:1,000, Affinity, USA) overnight at 4 °C.
CCK-8 assay
For CCK-8 assay, si-circRAD18 [5000] and si-control cells [5000] were placed in a 96-well plate. The plate was maintained for a certain time point at 37 °C, then CCK-8 solution (10 μL) was loaded, and the absorbance measured.
Measurement of glucose consumption and lactate production
An Amplex Red Glucose/Glucose Oxidase Assay Kit (Invitrogen, USA) was used to measure glucose consumption and lactate production. Based on the total cellular protein amounts, the data were normalized.
Transwell assay
Totally, 3×104 cells were added to the superior chambers (FBS-free), and medium with 20% FBS was loaded to the inferior chambers. The migrated PT cells were counted, then fixed with methanol and stained with 2% crystal violet using a migration chamber (BD Biosciences).
Luciferase reporter assay
Cells were placed in 96-well plates (5×103 cells) and the reporting plasmids (circRAD18-wt/mut or PDK1 3’-UTR-wt/mut) and inhibitors/mimics of certain miRNA were co-transfected using a Promega luciferase kit.
RNA immunoprecipitation (RIP)
Before RIP assays, cells were instantaneously transfected with MS2bs-circRAD18, MS2bs-circRAD18-mt, and MS2bs-Rluc. Moreover, RIP was performed with an RNA-Binding Protein Immunoprecipitation Kit, and miR-516b enrichment was determined after the purification. Ananti-Ago2 antibody (Millipore) was also used, and the relative amount of PDK1 mRNA, circRAD18, and miR-516b was checked.
Statistical analysis
All statistical analysis was performed using SPSS 25.0 software. Groups were compared by using Student’s t-test. All data were reported as the mean ± standard deviation (SD), and P<0.05 was regarded as statistically significant.
Results
circRAD18 is upregulated in papillary thyroid cancer with circular RNA structure
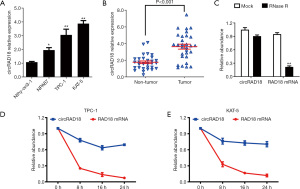
Compared to Nthy-ori3-1, circRAD18 was upregulated in three papillary thyroid carcinoma cell lines (Figure 1A). We then tested circRAD18 in 30 paired tissues by RT-qPCR analysis, and the data indicated that it was upregulated in papillary thyroid cancer samples (Figure 1B). The circular characteristics and degradation of circRAD18 by RNase R resistant assays and actinomycin D digestion assays was then examined and showed circRAD18 was resisted to RNase R, while the linear form RAD18 mRNA was degraded after exposure to RNase R (Figure 1C). Consistently, the circular structure of circRAD18 was more stable than the linear structure RAD18 mRNA in both KAT-5 and TPC-1cell lines (Figure 1D,1E).
Silence of circRAD18 inhibits the proliferation and glucose metabolism of papillary thyroid cancer cells
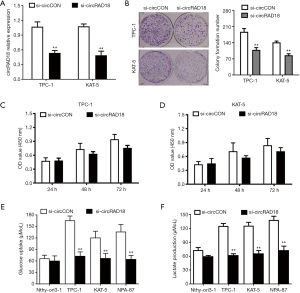
When examined by RT-qPCR analysis, the relative expression level of circRAD18 was decreased after being transfected with siRNA (Figure 2A). Colony-formation assays showed inhibition of circRAD18 suppressed the cell colony formatting ability of KAT-5 and TPC-1 cells (Figure 2B), while CCK-8 proliferation assays revealed downregulation of circRAD18 inhibited proliferation ability (Figure 2C,2D). Moreover, we found that while silence of circRAD18 could significantly suppress cell glucose uptake and lactate production in papillary thyroid cancer cells (Figure 2E,2F), it had no effect on normal cell lines.
Knockdown of circRAD18 suppresses the metastasis of papillary thyroid cancer cells
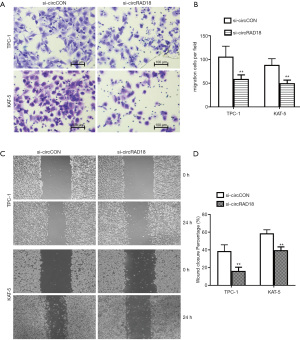
We next performed migration and invasion associated assays to further determine whether circRAD18 had a role in the metastasis of papillary thyroid cancer, and the results of transwell assays showed that silencing circRAD18 could attenuate the migration percentage of KAT-5 and TPC-1 cells (Figure 3A,3B). Moreover, inhibition of circRAD18 could suppress the migration ability of these cells in wound-healing experiments (Figure 3C,3D).
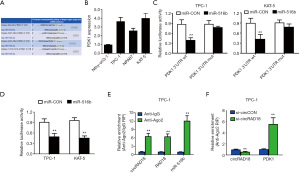
circRAD18 acts as a sponge of miR-516b in papillary thyroid cancer
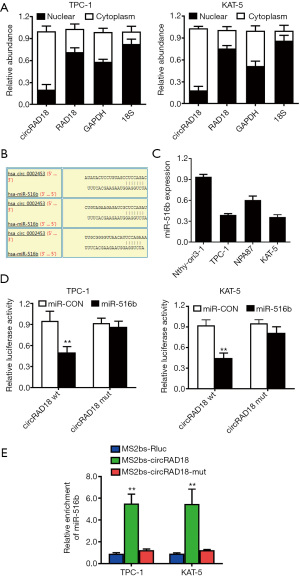
We determined the subcellular location of circRAD18 via qPCR analysis after isolating cytoplasmic and nuclear portions. The results showed circRAD18 predominantly existed in cytoplasm, which is where miRNA is mostly located, indicating interaction between the two (Figure 4A). Among the candidates, miR-516b was predicted to have the potential to bind circRAD18 sequence which contained three binding elements (Figure 4B). Moreover, we tested the relative expression of miR-516b by RT-qPCR analysis in cell lines and found miR-516b was downregulated (Figure 4C). Dual luciferase reporter assays also indicated that circRAD18 was able to interact with miR-516b (Figure 4D). To further validate this, we conducted MS2 related RNA immunoprecipitation (RIP) assays to confirm the direct interaction between circRAD18 and miR-516b, and the results revealed that miR-516b was extremely enriched in the MS2bs-circRAD18 group (Figure 4E).
circRAD18 regulates papillary thyroid cancer metabolism reprograming through the circRAD18-miR-516b-PDK1 axis
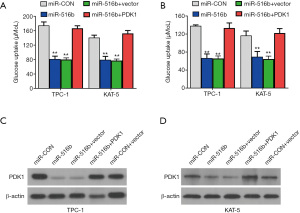
The TargetScan database was utilized to predict miR-516b targets and screen the downstream effects of miR-516b. Among the candidates (Figure 5A), PDK1, which is a metabolic protein in glucose intake (25) and is crucial for cancer progression (26,27), was identified as a target oncogene, and was overexpressed in papillary thyroid cancer cells (Figure 5B). Luciferase reporter experiments and RIP assays were also performed to verify whether miR-516b was able to directly bind the 3’-UTR of PDK1 mRNA. After transfecting miR-516b mimics, the luciferase amount was reduced (Figure 5C), and the expression level of PDK1 mRNA was decreased by miR-516b mimics, indicating that PDK1 is the target of miR-516b (Figure 5D). RISC complex of AGO2 protein related RIP assays were also conducted, and showed circRAD18, PDK1, and miR-516b were all gathered in the anti-AGO2 group (Figure 5E). Downregulation of circRAD18 resulted in significant upregulation of PDK1 gathering to RISC (Figure 5F), and transfection of miR-516b mimics could significantly suppress cell lactate production and glucose uptake (Figure 6A). Additionally, lactate production and glucose uptake returned to its previous level after overexpression of PDK1 (Figure 6B). Further, introduction of miR-516b mimics could decrease the protein expression of PDK1, which could be reversed by supplementation withPDK1 protein (Figure 6C,6D).
Discussion
CircRNAs, which are composed of non-coding RNAs, are recently discovered and have quickly become a hot spot in biomedical biotechnology (28,29). Previous studies have shown that the expression of circRNAs differs in not only cell types, but also in tissue types, and the differential expression level of many circRNAs can serve as potential biomarkers to aid in the diagnosis and treatment of different cancers (30,31). Scientists have gradually come to realize that these novel RNAs with circular characteristics are not unimportant byproducts of splicing, but are vital modulators of a variety of cellular processes (32) and are involved in tumor development and progression. By translating the novel 370 amino acid CTNNB1 subtype, CTNNB1 pre-mRNA derived circRNAs promoted HCC cell proliferation by activating the Wnt signaling pathway (33). By sponging two miRNAs, miR-589 and miR-545, circular RNA PRKCI was upregulated in lung adenocarcinoma and promoted tumorigenesis (34), and in triple-negative breast cancer, circRNAs were discovered as tumor promotors by reducing apoptosis and promoting proliferation (35,36). Nevertheless, the role and function of circRNAs in papillary thyroid carcinoma has been rarely studied. In one report, CIRC_0006156 was upregulated in serum exosomes that promote the progression of papillary thyroid carcinoma via the miR-1178/TLR4 axis (37), and in another, CircRAPGF5 was seen to regulate the progression of papillary thyroid carcinoma (38). CIRC0088494 was also seen to be upregulated in papillary thyroid carcinoma and promoted carcinogenesis via the miR-876-CCND1 axis (39).
Our results identified circRAD18 as a frequently upregulated novel circRNA in papillary thyroid cancer, and knockdown of circRAD18 significantly inhibited its growth and migration ability. Additionally, we found that circRAD18 was involved in glucose metabolism reprogramming of papillary thyroid cancer. Silence of circRAD18 remarkably inhibited cell glucose uptake and lactate production in papillary thyroid cancer cells, suggesting that it could directly block miR-516b to promote thyroid papillary carcinoma progression by stimulating the expression of glucose metabolism protein PDK1.
Previous research has shown that miR-516b was downregulated and an important tumor suppressor in primary melanoma (40), and in another study, highly upregulated circRNA, circCDC45 promoted glioma cell proliferation and metastasis by blocking miR-516b (41). Linc-01123 was also seen to regulate the Hedgehog pathway by the miR-516b-5p/Gli1 axis in osteosarcoma (42). In the recent decades, herbs and extractions from TCM, are gaining acceptance as promising complementary and alternative medicines for numerous diseases treatment (43-46). As a proven target of miR-516b, PDK1 regulates glucose and fatty acid metabolism and homeostasis (26,27). According to our findings, PDK1 was validated to be the downstream gene of miR-516b in papillary thyroid cancer, and circRAD18 could enhance the expression of PDK1.
In conclusion, our study illustrated the biological functions of circRAD18 in papillary thyroid carcinoma, and revealed the function of the circRAD18-miR-516b-PDK1 axis in papillary thyroid cancer metabolic reprogramming.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 81772961).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/gs-21-481
Data Sharing Statement: Available at https://dx.doi.org/10.21037/gs-21-481
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/gs-21-481). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The Ethics Committee of SYSUCC approved the study (GZR2017163). Informed consent was obtained from all involved patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Santiago K, Chen Wongworawat Y, Khan S. Differential MicroRNA-Signatures in Thyroid Cancer Subtypes. J Oncol 2020;2020:2052396 [Crossref] [PubMed]
- Park H, Park J, Park SY, et al. Clinical Course from Diagnosis to Death in Patients with Well-Differentiated Thyroid Cancer. Cancers (Basel) 2020;12:2323. [Crossref] [PubMed]
- Sakai T, Sugitani I, Ebina A, et al. Active Surveillance for T1bN0M0 Papillary Thyroid Carcinoma. Thyroid 2019;29:59-63. [Crossref] [PubMed]
- Pelizzo MR, Merante Boschin I, Toniato A, et al. Diagnosis, treatment, prognostic factors and long-term outcome in papillary thyroid carcinoma. Minerva Endocrinol 2008;33:359-79. [PubMed]
- Saravana-Bawan B, Bajwa A, Paterson J, et al. Active surveillance of low-risk papillary thyroid cancer: A meta-analysis. Surgery 2020;167:46-55. [Crossref] [PubMed]
- Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol 2015;12:381-8. [Crossref] [PubMed]
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141-57. [Crossref] [PubMed]
- Wang Y, Hou J, He D, et al. The Emerging Function and Mechanism of ceRNAs in Cancer. Trends Genet 2016;32:211-24. [Crossref] [PubMed]
- Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res 2013;73:5609-12. [Crossref] [PubMed]
- Shan K, Liu C, Liu BH, et al. Circular Noncoding RNA HIPK3 Mediates Retinal Vascular Dysfunction in Diabetes Mellitus. Circulation 2017;136:1629-42. [Crossref] [PubMed]
- Devaux Y, Creemers EE, Boon RA, et al. Circular RNAs in heart failure. Eur J Heart Fail 2017;19:701-9. [Crossref] [PubMed]
- Huang JL, Qin MC, Zhou Y, et al. Comprehensive analysis of differentially expressed profiles of Alzheimer's disease associated circular RNAs in an Alzheimer's disease mouse model. Aging (Albany NY) 2018;10:253-65. [Crossref] [PubMed]
- Yu T, Wang Y, Fan Y, et al. CircRNAs in cancer metabolism: a review. J Hematol Oncol 2019;12:90. [Crossref] [PubMed]
- Xiao W, Zheng S, Zou Y, et al. CircAHNAK1 inhibits proliferation and metastasis of triple-negative breast cancer by modulating miR-421 and RASA1. Aging (Albany NY) 2019;11:12043-56. [Crossref] [PubMed]
- Yang W, Yang X, Wang X, et al. Silencing CDR1as enhances the sensitivity of breast cancer cells to drug resistance by acting as a miR-7 sponge to down-regulate REGγ. J Cell Mol Med 2019;23:4921-32. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Weng W, Wei Q, Toden S, et al. Circular RNA ciRS-7-A Promising Prognostic Biomarker and a Potential Therapeutic Target in Colorectal Cancer. Clin Cancer Res 2017;23:3918-28. [Crossref] [PubMed]
- Zou Y, Zheng S, Deng X, et al. Diagnostic and prognostic value of circular RNA CDR1as/ciRS-7 for solid tumours: A systematic review and meta-analysis. J Cell Mol Med 2020;24:9507-17. [Crossref] [PubMed]
- Zou Y, Zheng S, Deng X, et al. The Role of Circular RNA CDR1as/ciRS-7 in Regulating Tumor Microenvironment: A Pan-Cancer Analysis. Biomolecules 2019;9:429. [Crossref] [PubMed]
- Yang Y, Gao X, Zhang M, et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J Natl Cancer Inst 2018; [Crossref] [PubMed]
- Ye F, Gao G, Zou Y, et al. circFBXW7 Inhibits Malignant Progression by Sponging miR-197-3p and Encoding a 185-aa Protein in Triple-Negative Breast Cancer. Mol Ther Nucleic Acids 2019;18:88-98. [Crossref] [PubMed]
- Zou Y, Zheng S, Xiao W, et al. circRAD18 sponges miR-208a/3164 to promote triple-negative breast cancer progression through regulating IGF1 and FGF2 expression. Carcinogenesis 2019;40:1469-79. [Crossref] [PubMed]
- Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis 2018;9:417. [Crossref] [PubMed]
- Xu X, Chen Y, Fu Q, et al. The chemical diversity and structure-based discovery of allosteric modulators for the PIF-pocket of protein kinase PDK1. J Enzyme Inhib Med Chem 2019;34:361-74. [Crossref] [PubMed]
- Atas E, Oberhuber M, Kenner L. The Implications of PDK1-4 on Tumor Energy Metabolism, Aggressiveness and Therapy Resistance. Front Oncol 2020;10:583217 [Crossref] [PubMed]
- Gagliardi PA, Puliafito A, Primo L. PDK1: At the crossroad of cancer signaling pathways. Semin Cancer Biol 2018;48:27-35. [Crossref] [PubMed]
- Li S, Teng S, Xu J, et al. Microarray is an efficient tool for circRNA profiling. Brief Bioinform 2019;20:1420-33. [Crossref] [PubMed]
- Vo JN, Cieslik M, Zhang Y, et al. The Landscape of Circular RNA in Cancer. Cell 2019;176:869-881.e13. [Crossref] [PubMed]
- Shi Y, Song R, Wang Z, et al. Potential clinical value of circular RNAs as peripheral biomarkers for the diagnosis and treatment of major depressive disorder. EBioMedicine 2021;66:103337 [Crossref] [PubMed]
- Chen X, Gu M, Zhang X. Circular RNAs in compression-induced intervertebral disk degeneration. EBioMedicine 2020;54:102720 [Crossref] [PubMed]
- Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 2016;17:205-11. [Crossref] [PubMed]
- Liang WC, Wong CW, Liang PP, et al. Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol 2019;20:84. [Crossref] [PubMed]
- Qiu M, Xia W, Chen R, et al. The Circular RNA circPRKCI Promotes Tumor Growth in Lung Adenocarcinoma. Cancer Res 2018;78:2839-51. [Crossref] [PubMed]
- Kong Y, Yang L, Wei W, et al. CircPLK1 sponges miR-296-5p to facilitate triple-negative breast cancer progression. Epigenomics 2019;11:1163-76. [Crossref] [PubMed]
- Liu P, Zou Y, Li X, et al. circGNB1 Facilitates Triple-Negative Breast Cancer Progression by Regulating miR-141-5p-IGF1R Axis. Front Genet 2020;11:193. [Crossref] [PubMed]
- Wu G, Zhou W, Pan X, et al. Circular RNA Profiling Reveals Exosomal circ_0006156 as a Novel Biomarker in Papillary Thyroid Cancer. Mol Ther Nucleic Acids 2020;19:1134-44. [Crossref] [PubMed]
- Liu W, Zhao J, Jin M, et al. circRAPGEF5 Contributes to Papillary Thyroid Proliferation and Metastatis by Regulation miR-198/FGFR1. Mol Ther Nucleic Acids 2019;14:609-16. [Crossref] [PubMed]
- Lou W, Ding B, Wang J, et al. The Involvement of the hsa_circ_0088494-miR-876-3p-CTNNB1/CCND1 Axis in Carcinogenesis and Progression of Papillary Thyroid Carcinoma. Front Cell Dev Biol 2020;8:605940 [Crossref] [PubMed]
- Hanniford D, Segura MF, Zhong J, et al. Identification of metastasis-suppressive microRNAs in primary melanoma. J Natl Cancer Inst 2015;107:dju494 [Crossref] [PubMed]
- Liu J, Hou K, Ji H, et al. Overexpression of circular RNA circ-CDC45 facilitates glioma cell progression by sponging miR-516b and miR-527 and predicts an adverse prognosis. J Cell Biochem 2020;121:690-7. [Crossref] [PubMed]
- Pan X, Tan J, Tao T, et al. LINC01123 enhances osteosarcoma cell growth by activating the Hedgehog pathway via the miR-516b-5p/Gli1 axis. Cancer Sci 2021;112:2260-71. [Crossref] [PubMed]
- Peng F, Xiong L, Peng C. (-)-Sativan Inhibits Tumor Development and Regulates miR-200c/PD-L1 in Triple Negative Breast Cancer Cells. Front Pharmacol 2020;11:251. [Crossref] [PubMed]
- Peng F, Tang H, Du J, et al. Isoliquiritigenin Suppresses EMT-Induced Metastasis in Triple-Negative Breast Cancer through miR-200c/C-JUN/Formula: see text-Catenin. Am J Chin Med 2021;49:505-23. [Crossref] [PubMed]
- Peng F, Xiong L, Xie X, et al. Isoliquiritigenin Derivative Regulates miR-374a/BAX Axis to Suppress Triple-Negative Breast Cancer Tumorigenesis and Development. Front Pharmacol 2020;11:378. [Crossref] [PubMed]
- Wang L, Peng F, Peng C, et al. Gut Microbiota in Tumor Microenvironment: A Critical Regulator in Cancer Initiation and Development as Potential Targets for Chinese Medicine. Am J Chin Med 2021;49:609-26. [Crossref] [PubMed]
(English Language Editor: B. Draper)


