
The relationship between autophagy-related genes and the staging and prognosis of thyroid cancer: a bioinformatics analysis
Introduction
Thyroid cancer is a malignant tumor originating from thyroid follicular epithelium or para-follicular epithelial cells (1). It is also the most common malignant tumor of the head and neck. Thyroid cancer can be pathologically divided into papillary thyroid carcinoma, follicular thyroid carcinoma, medullary thyroid carcinoma, and anaplastic thyroid carcinoma (1). Among these thyroid cancers, papillary carcinoma is the most common, and accounts for about 80% of all thyroid cancers (2). Papillary carcinoma often occurs in young women, is highly differentiated, and has a good prognosis (2). Thyroid follicular carcinoma accounts for about 10% of thyroid cancers. It is more common in middle-aged women, is moderately differentiated, and has a relatively poor prognosis (3). The degree of malignancy of undifferentiated thyroid cancer is extremely high, the survival time is only 7–10 months, and the prognosis is poor (3). The prognosis of medullary thyroid cancer lies between differentiated thyroid cancer and undifferentiated thyroid cancer (3). Deaths from thyroid cancer mainly occur in patients aged 70 years or older, and are increasing year by year (4,5). Thyroid cancer ranks as the 17th most common malignant tumor in men and the 5th most common malignant tumor in women (6). Gene mutation is an important feature of malignant tumors, which can affect the occurrence and prognosis of tumors (7). At present, the v-raf murine sarcoma viral oncogene homolog B1 (BRAF) gene and Rat sarcoma (RAS) genes are the most researched genes in thyroid cancer. Notably, research has shown that when the codon 600 of BRAF gene is mutated, it is one of the most aggressive phenotypes in papillary thyroid cancer (8).
In recent years, the relationship between autophagy-related genes and tumors has received attention. Autophagy refers to the responses of cells to changes in internal and external environmental pressures. It is a mechanism that exists in organisms to purify their own redundant or damaged organelles during their development and aging (9,10). Autophagy generally refers to macroautophagy and can be divided into the following 3 types: macroautophagy, small autophagy, and molecular chaperone-mediated autophagy (11,12). When apoptosis is inhibited, autophagy plays a role in promoting cell death (13,14). Autophagy has the dual effects of promoting and inhibiting the occurrence and development of tumors, and its specific mechanism is not completely clear. More studies need to be conducted to confirm whether autophagy can be used as a new target for tumor therapy (15,16). At present, some studies have shown that autophagy is closely related to thyroid cancer (17,18). However, differences in the expression of autophagy-related genes in thyroid cancer and their relationship with prognosis remains unclear. This study focused on the profile or landscape of autophagy-related genes in thyroid cancer tissues, and analyzed the relationship between autophagy-related genes and the prognosis of patients with thyroid cancer. We present the following article in accordance with the REMARK reporting checklist (available at https://dx.doi.org/10.21037/gs-21-480).
Methods
Research object and data source
The standardized ribonucleic acid (RNA) sequencing data and corresponding clinical records of 497 patients were obtained from The Cancer Genome Atlas (TCGA) database loaded on cbioportal.org (19). In the original database, detailed clinical characteristics are recorded, including data on age, gender, tumor grade, pathological information, and laboratory test results. The diagnosis of thyroid cancer is based on the results of pathological examinations. The gene expression level is shown as the z-score of the messenger RNA (mRNA), and is compared between each subject. These data sets are publicly available, and have been exempted from ethical approval by the Ethics Committee of our hospital. Patients signed informed consent forms. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Bioinformatics analysis
Similar to other studies, in this study, a collection of autophagy-related genes was acquired from rectom.org (identification: R-HSA-1632852). Based on the completeness of the sequencing and prognostic data, 135 effective genes were screened to form a gene set (20). To distinguish between samples based on gene expression profiles, a cluster analysis was performed to examine the genetic expression of the entire genome. We identified cases with similar gene expression patterns from the entire study population. The transcription levels of related genes are shown as mRNA z-scores, and were clustered by the Stanford program using a hierarchical clustering algorithm, as described previously (21). We used the Java Treeview program (jtreeview.sourceforge.net) (22) and GraphPad Prism (version 8.0, GraphPad Software, Inc., San Diego, California, USA) to generate cluster heat maps and patterns for specific tumor stages.
Prognostic correlation analysis
We compared the survival expression levels of autophagy-related genes in different groups to study their prognostic effects. The 4 main outcomes were as follows: overall survival (OS), progression-free survival (PFS), disease-free survival (DFS), and disease-specificity survival (DSS). These results were analyzed using GraphPad Prism (version 8.0, GraphPad Software, Inc., San Diego, California, USA). We compared the survival rates of different clusters to examine the relationship between related gene expression levels and prognosis. In addition, GraphPad Prism (version 8.0, GraphPad Software, Inc., San Diego, California, USA) was used to analyze the OS differences between cohorts with low or high expression levels of specific genes.
Statistical analysis
SPSS 24.0 (IBM, NY, USA) was used for the statistical analysis. Continuous variables are expressed as mean ± standard deviation (mean ± SD). Categorical variables are represented by numbers and were compared using χ2 test or Fisher’s exact test. An analysis of variance was used to detect differences in gene expression levels between clusters. The correlations between the variables were determined by regression analyses. The survival curves of different groups were drawn and compared using the log-rank test in GraphPad Prism (version 8.0, GraphPad Software, Inc., CA, USA). A P value <0.05 was considered statistically significant.
Results
Autophagy-related gene expression profile is related to the prognosis of thyroid cancer
Based on the hierarchical clustering, the 497 patients were divided into 2 clusters and 4 subclusters (see Figure 1A and Table 1). A comparison of the clinical parameters of the 2 clusters showed that there were differences in node (N)-stage, and a comparison of the 4 subclusters showed that there were differences in age and 4 other characteristics. In relation to the survival comparison, there was a difference in DFS between the 2 clusters, and there was a difference in OS and DFS between the subclusters (see Figure 1B,1C, and Table 2).
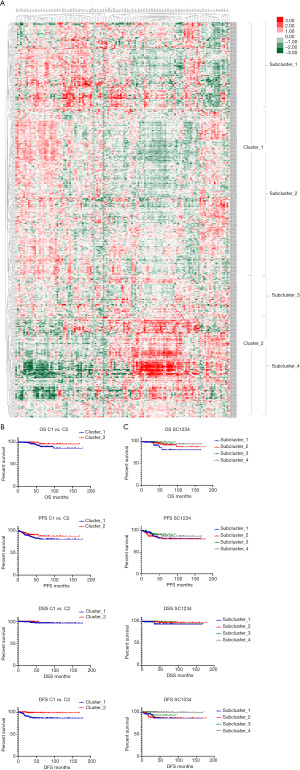
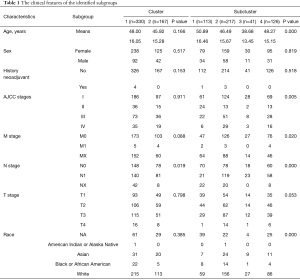
Full table
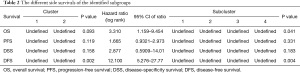
Full table
Genes with different expressions
A comparison of the differentially expressed genes (DEGs) showed that the 2 clusters had 114 DEGs, and the 4 subclusters had 131 DEGs (see Table S1). Figure 2A and Figure 2B respectively list the most significant genes.
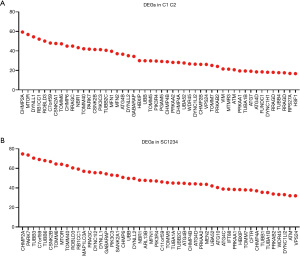
The relationship between autophagy-related gene expression and various factors
In relation to the 5 different factors in each group, there was no statistical difference between N0N1NX in Clusters 1 and 2 (P=0.19), and the distribution of the subclusters was statistically different (P<0.001; see Figure 3, Table S2). Additionally, the distribution of M0M1MX in the subclusters was statistically different (see Figure 4, Table S3). The distribution of age in the subclusters was not statistically different (P=0.901; see Figure 5A); however, the distribution of the American Joint Committee on Cancer (AJCC) stage in the subclusters was statistically different (P=0.005; see Figure 5B). The genes related to age and AJCC stage are shown in Figure 5C,5D.
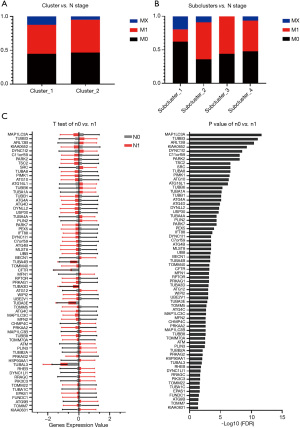
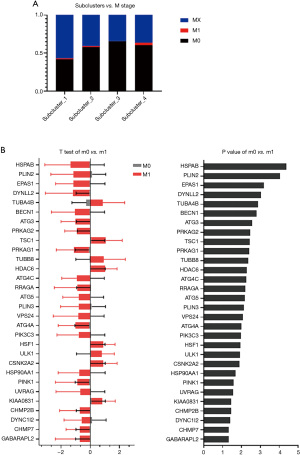
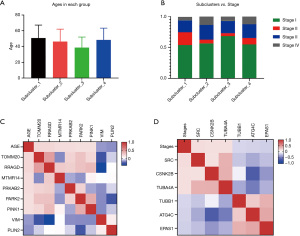
The relationship between clinical staging and differential genes
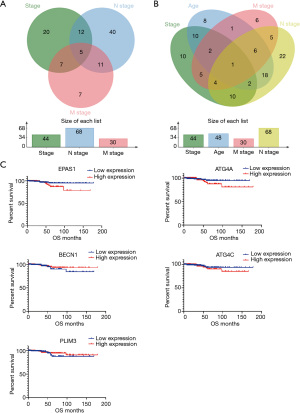
The clinical indicators stage/N stage/ metastasis (M) stage and age-related DEGs are listed in Table 3. In relation to the stage/N stage/M stage-related DEGs, 5 common genes were identified: EPAS1, ATG4A, BECN1, ATG4C, and PLIN3 (Figure 6A). Additionally, in relation to the stage/N stage/M stage-related DEGs and age-related DEGs 1 common gene was identified: EPAS1 (see Figure 6B). The correlations between the 5 common genes and OS are listed in Figure 6C. The results of the statistical tests and hazard ratios (HRs) are listed in Table 4.

Full table
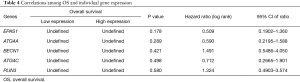
Full table
Discussion
This study showed that certain clinical features are closely related to the prognosis of thyroid cancer, and gene expression patterns are related to autophagy. Statistical differences were found in relation to the N stage between the different clusters, and differences were also found in relation to age, the AJCC stage, M stage, N stage, and race among the subclusters. In terms of prognosis, there were differences in DFS between the 2 clusters. The long-term DFS rate of Cluster 2 was higher than that of Cluster 1. The OS and DFS rates were different among the 4 subclusters. There were many DEGs in the different sets. After analyzing some factors, the following 5 shared DEGs were identified in the differential genes grouped by tumor stage: EPAS1, ATG4A, BECN1, ATG4C, and PLIN3. These results indicate that these genes are closely related to the staging of thyroid cancer. After examining the differential genes across different ages, only 1 DEG (i.e., EPAS1) was identified. However, there does not seem to be a clear relationship between the genes whose expressions were different due to different factors and the prognosis of patients with thyroid cancer.
There has been an increasing trend in the incidence of thyroid cancer in recent years (3). Studies have shown that this increase is related to the timely diagnosis and close monitoring brought about by advancements in medical technology, such that cases that were not detected in time previously are now being discovered (3). Thyroid cancer has a variety of clinical features from indolent tumors with low mortality in most cases to very aggressive malignancies (such as anaplastic thyroid cancer). Thus, the main challenge doctors face is to identify high-risk patients and perform appropriate diagnostic tests to choose the most effective treatment plan (23). The prognosis of thyroid cancer also has a large heterogeneity, even regionally; however, most patients have a good prognosis (24,25). Many studies have examined the prognosis of the thyroid gland; for example, studies have been conducted on the BRAF gene mutation, RAS gene mutation, rearranged during transfection gene rearrangement, telomerase reverse transcriptase promoter mutation, and phosphatase and TENsin gene mutation. However, relatively few studies have been conducted on the relationship between these factors and tumor staging and their effects on long-term prognosis (26-28).
Hypoxia has been reported to be involved in multiple pathways regulating tumor cells (29). EPAS1 is a protein family member with a basic Helixdoop-helix/PAS structure, and it is a key hypoxia-related transcription factor related to tumor progression (30-32). Some research has shown that the expression of EPAS1 in normal tissues of the body is low or has no expression; however, it is abnormally high in malignant tumor tissues, and it is involved in a series of biological behaviors of cancer cells (33). Studies have also shown that EPAS1 plays a vital role in the pathogenesis of esophageal squamous cell carcinoma and may be used as a prognostic marker and therapeutic target (34). The present study showed that EPAS1 differed significantly in the AJCC stages/N stage/M stages, and in different ages, and is the only 1 gene with differential expression. Thus, EPAS1 may play an important role in the occurrence and development of thyroid cancer.
Putra et al. found that the genetic polymorphism of the EPAS1 gene may lead to changes in its gene expression level, thereby driving the development of cancer and becoming a prognostic indicator of non-small cell lung cancer (35). Mohammed et al. showed that plasma EPAS1 mRNA levels may be an indicator of poor prognosis for patients with advanced colorectal cancer. They also found that high levels of EPAS1 in plasma are associated with being aged over 50 years, disease recurrence, and patient mortality. When patients were divided into early (I and II) and late (III and IV) groups, correlations were observed between high levels of EPAS1 mRNA and poor DFS and late OS (36). In this study, we found that EPAS1 expression levels differed in TC patients of different stages and ages, but in the analysis of the relationship with long-term prognosis, no differences in the survival of patients with different expression levels of EPAS1 were found; however, this may be due to the sample size of the study.
This study had a number of limitations. First, a retrospective bioinformatics analysis was conducted. The TCGA database provides detailed clinical data, but the sample size was relatively small for the analysis of survival rates. There are often many prognostic-related factors. However the results of this study only showed some genes expressed differences in different groups, and no statistically significant relationships between the genes and long-term prognosis was found. In the future, prospective observational studies should be carried out to study specific genes to observe the effects of these genes on treatment responses and their relationships with the long-term prognosis of patients with thyroid cancer.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://dx.doi.org/10.21037/gs-21-480
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/gs-21-480). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. These data sets are publicly available, and have been exempted from ethical approval by the Ethics Committee of our hospital. Patients signed informed consent forms. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Asa SL. The Current Histologic Classification of Thyroid Cancer. Endocrinol Metab Clin North Am 2019;48:1-22. [Crossref] [PubMed]
- Janjua N, Wreesmann VB. Aggressive differentiated thyroid cancer. Eur J Surg Oncol 2018;44:367-77. [Crossref] [PubMed]
- Seib CD, Sosa JA. Evolving Understanding of the Epidemiology of Thyroid Cancer. Endocrinol Metab Clin North Am 2019;48:23-35. [Crossref] [PubMed]
- Azadnajafabad S, Saeedi Moghaddam S, Mohammadi E, et al. Global, regional, and national burden and quality of care index (QCI) of thyroid cancer: A systematic analysis of the Global Burden of Disease Study 1990-2017. Cancer Med 2021;10:2496-508. [Crossref] [PubMed]
- Deng Y, Li H, Wang M, et al. Global Burden of Thyroid Cancer From 1990 to 2017. JAMA Netw Open 2020;3:e208759 [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Nikiforov YE, Nikiforova MN. Molecular genetics and diagnosis of thyroid cancer. Nat Rev Endocrinol 2011;7:569-80. [Crossref] [PubMed]
- Xing M. BRAF mutation in thyroid cancer. Endocr Relat Cancer 2005;12:245-62. [Crossref] [PubMed]
- Poillet-Perez L, Xie X, Zhan L, et al. Autophagy maintains tumour growth through circulating arginine. Nature 2018;563:569-73. [Crossref] [PubMed]
- Katheder NS, Khezri R, O'Farrell F, et al. Microenvironmental autophagy promotes tumour growth. Nature 2017;541:417-20. [Crossref] [PubMed]
- Maiuri MC, Zalckvar E, Kimchi A, et al. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol 2007;8:741-52. [Crossref] [PubMed]
- Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol 2010;221:3-12. [Crossref] [PubMed]
- Yoshida GJ. Therapeutic strategies of drug repositioning targeting autophagy to induce cancer cell death: from pathophysiology to treatment. J Hematol Oncol 2017;10:67. [Crossref] [PubMed]
- D'Arcy MS. Cell death: a review of the major forms of apoptosis, necrosis and autophagy. Cell Biol Int 2019;43:582-92. [Crossref] [PubMed]
- Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer 2017;17:528-42. [Crossref] [PubMed]
- Amaravadi RK, Kimmelman AC, Debnath J. Targeting Autophagy in Cancer: Recent Advances and Future Directions. Cancer Discov 2019;9:1167-81. [Crossref] [PubMed]
- Kim HM, Kim ES, Koo JS. Expression of Autophagy-Related Proteins in Different Types of Thyroid Cancer. Int J Mol Sci 2017;18:540. [Crossref] [PubMed]
- Plantinga TS, Tesselaar MH, Morreau H, et al. Autophagy activity is associated with membranous sodium iodide symporter expression and clinical response to radioiodine therapy in non-medullary thyroid cancer. Autophagy 2016;12:1195-205. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Türei D, Földvári-Nagy L, Fazekas D, et al. Autophagy Regulatory Network - a systems-level bioinformatics resource for studying the mechanism and regulation of autophagy. Autophagy 2015;11:155-65. [Crossref] [PubMed]
- de Hoon MJ, Imoto S, Nolan J, et al. Open source clustering software. Bioinformatics 2004;20:1453-4. [Crossref] [PubMed]
- Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics 2004;20:3246-8. [Crossref] [PubMed]
- Cabanillas ME, McFadden DG, Durante C. Thyroid cancer. Lancet 2016;388:2783-95. [Crossref] [PubMed]
- Huang M, Yan C, Wei H, et al. Clinicopathological characteristics and prognosis of thyroid cancer in northwest China: A population-based retrospective study of 2490 patients. Thorac Cancer 2018;9:1453-60. [Crossref] [PubMed]
- Kim HI, Kim TH, Choe JH, et al. Restratification of survival prognosis of N1b papillary thyroid cancer by lateral lymph node ratio and largest lymph node size. Cancer Med 2017;6:2244-51. [Crossref] [PubMed]
- Abdullah MI, Junit SM, Ng KL, et al. Papillary Thyroid Cancer: Genetic Alterations and Molecular Biomarker Investigations. Int J Med Sci 2019;16:450-60. [Crossref] [PubMed]
- Brose MS, Cabanillas ME, Cohen EE, et al. Vemurafenib in patients with BRAF(V600E)-positive metastatic or unresectable papillary thyroid cancer refractory to radioactive iodine: a non-randomised, multicentre, open-label, phase 2 trial. Lancet Oncol 2016;17:1272-82. [Crossref] [PubMed]
- Howell GM, Hodak SP, Yip L. RAS mutations in thyroid cancer. Oncologist 2013;18:926-32. [Crossref] [PubMed]
- Shen J, Xia W, Khotskaya YB, et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature 2013;497:383-7. [Crossref] [PubMed]
- Wu D, Rastinejad F. Structural characterization of mammalian bHLH-PAS transcription factors. Curr Opin Struct Biol 2017;43:1-9. [Crossref] [PubMed]
- Ryu JH, Yang S, Shin Y, et al. Interleukin-6 plays an essential role in hypoxia-inducible factor 2α-induced experimental osteoarthritic cartilage destruction in mice. Arthritis Rheum 2011;63:2732-43. [Crossref] [PubMed]
- Hagstrom SA, Ying GS, Pauer GJ, et al. Endothelial PAS domain-containing protein 1 (EPAS1) gene polymorphisms and response to anti-VEGF therapy in the comparison of AMD treatments trials (CATT). Ophthalmology 2014;121:1663-4.e1. [Crossref] [PubMed]
- Xu XH, Bao Y, Wang X, et al. Hypoxic-stabilized EPAS1 proteins transactivate DNMT1 and cause promoter hypermethylation and transcription inhibition of EPAS1 in non-small cell lung cancer. FASEB J 2018; Epub ahead of print. [Crossref] [PubMed]
- Islam F, Gopalan V, Law S, et al. Molecular Deregulation of EPAS1 in the Pathogenesis of Esophageal Squamous Cell Carcinoma. Front Oncol 2020;10:1534. [Crossref] [PubMed]
- Putra AC, Eguchi H, Lee KL, et al. The A Allele at rs13419896 of EPAS1 Is Associated with Enhanced Expression and Poor Prognosis for Non-Small Cell Lung Cancer. PLoS One 2015;10:e0134496 [Crossref] [PubMed]
- Mohammed N, Rodriguez M, Garcia V, et al. EPAS1 mRNA in plasma from colorectal cancer patients is associated with poor outcome in advanced stages. Oncol Lett 2011;2:719-24. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


