
Seven-gene signature on tumor microenvironment for predicting the prognosis of patients with pancreatic cancer
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is a highly malignant neoplasm with a poor prognosis and high mortality; its overall 5-year survival rate is reported to be <7% (1). In the USA, PDAC has become the fourth leading cause of cancer-related death, and more than 45,000 Americans died of PDAC in 2019 (1). The absence of early detection tests and recognizable symptoms leads to only 15–20% of cases being diagnosed in the early resectable stages (2). The median overall survival for patients treated with pancreaticoduodenectomy in combination with adjuvant therapy is only approximately 35.0–54.4 months (3). Therefore, there is an urgent need to make a breakthrough in the research of pancreatic cancer in terms of early diagnosis and treatment. In addition to the progress in surgical resection and other adjuvant therapies, more specific and sensitive biomarkers for the prediction early diagnosis and prognosis can help in the development of effective therapeutic strategies.
Tumors are complex environments composed of endothelial cells, mesenchymal cells, immune cells, inflammatory mediators, and extracellular matrix molecules, known as the tumor microenvironment (TME) (4-7). Immune and stromal cells are 2 main components of the TME, and have been reported to be significant for the diagnostic and prognostic assessments of tumors (8-10). Yoshihara et al. developed an algorithm to predict the abundance of immune and stromal cells based on the gene expression data from The Cancer Genome Atlas (TCGA) database, which is called Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data (ESTIMATE) (11). Subsequently, the algorithm was quickly applied to a variety of tumor-related analyses, such as colon cancer (12), breast cancer (13), and glioblastoma (14), demonstrating the broad utility of the ESTIMATE algorithm. Nevertheless, the ESTIMATE algorithm has not been previously investigated in pancreatic cancer.
There have been many studies focused on the prognosis of PDAC (15,16). Some scholars have proposed risk models as to predict the long-term survival status of patients, and optimize the choice of treatment modalities. However, previous researches have mainly focused on the tumor itself, and few have focused on the TME. Our study took this perspective and investigated the gene changes related to microenvironment. In our study, by using TCGA database of pancreatic cancer, we identified a list of significantly differentially expressed genes (DEGs) with the prognostic significance relying on ESTIMATE algorithm-derived immune and stromal scores. Importantly, a reliable prognostic signature for PDAC patients was set up and validated in the Gene Expression Omnibus (GEO) database.
We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/gs-21-28).
Methods
Data and sources
Gene expression profiles (level 3, RNA sequencing) and clinical information of PDAC were obtained from TCGA (https://cancergenome.nih.gov/) up to July 1, 2019. Three microarray gene expression arrays and related clinical information of PDAC datasets (GSE78229, GSE62452, and GSE28735) were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/gds/). Non-tumor cases were discarded, and the remaining cases were used as the validation cohort. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
The ESTIMATE algorithm can calculate the percent abundance of immune and stromal cells (known as immune and stromal scores) via gene expression profiles in complex tissues (11). TCGA datasets were analyzed by the ESTIMATE algorithm, and immune and stromal scores were obtained. The workflow of this experiment is shown in Figure 1.
Identification of DEGs
The differential analysis was performed using the limma R package (version: 3.36.5; http://www.bioconductor.org/packages/release/bioc/html/limma.html) [|log2 fold-change (FC)| >1, false discovery rate (FDR) <0.05]. First, TCGA datasets were divided into a high immune score subgroup and low immune score subgroup, according to the median value. The DEG analysis was performed, and we obtained a series of immune-related DEGs. We repeated the procedure and obtained another set of stromal-related DEGs.
Volcano plot and heatmap
Volcano plots and heatmaps were generated by the GGPLOT (Version: 3.3.3; https://cran.r-project.org/web/packages/ggplot2/index.html) and pheatmap (Version: 1.0.12; https://cran.r-project.org/web/packages/pheatmap/index.html) R packages, respectively (17,18).
Venn diagram
A Venn diagram was generated by an open source web tool Venny 2.1 (https://bioinfogp.cnb.csic.es/tools/venny) with upregulated/downregulated genes from the immune and stromal subgroups (19).
Gene co-expression analysis
Pearson’s correlation coefficients between core genes of the risk model and DEGs were calculated with the limma R package, and the cut-off criteria were set as follows: correlation coefficients ≥0.6 and P<0.05.
Pathway enrichment analysis
Pathway enrichment analysis, including Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses, were implemented by the cluster Profiler R package (Version: 3.18.0; http://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html) (20). The cut-off criterion was set as P<0.05.
Protein–protein interaction (PPI) network and module analyses
The PPI analysis was performed by the STRING database (Version: 11.0; https://string-db.org) with DEGs (21). A confidence score ≥0.4 was used as the critical value, and Cytoscape (Version: 3.7.1; https://cytoscape.org) was used for further analysis. Molecular complex detection (MCODE) was then used to detect hub clustering modules in the PPI network (22).
Survival analysis and receiver-operating characteristic (ROC) curve
To eliminate the interference of surgical blow to the overall prognosis of patients, cases with overall survival <10 days were excluded from our subsequent analysis. Univariate Cox analysis was performed with R package survival by setting a cut-off point of P<0.05. Multivariate Cox regression analysis was also implemented by R package survival. Subsequently, we constructed a prognostic risk score model with the following formula: risk score = expression of gene 1 × β1 + expression of gene 2 × β2 +…+ expression of gene n × βn, where β is the regression coefficient. The risk score of each patient was then calculated, and cases were grouped into high- and low-risk subgroups according to the median values. Predictive accuracy and sensitivity were evaluated by ROC curve analysis with R package survival ROC.
Statistical analysis
All statistical analyses were performed by R software (version 3.5.3). The χ2-test was used to evaluate the association between the risk score and clinical characteristics. Univariate and multivariate Cox regression models were used to analyze prognostic factors. P<0.05 was considered statistically significant.
Results
Comparison of gene expression profile with immune and stromal scores in PDAC
Based on the ESTIMATE algorithm, we obtained immune and stromal scores from TCGA database of pancreatic cancer. As shown in Figure 2A, the median value of immune scores was 1,170.63 (range, –602.44 to 2,662.36), while the median value of stromal scores was 787.21 (range, –1,658.32 to 2,171.03). To find out the potential correlation of gene expression profiles with immune scores and/or stromal scores, we divided 178 PDAC cases into top and bottom halves (high- vs. low-score groups) based on the median value of the scores. We then conducted limma analysis to select DEGs using gene expression data, depending on differences of scores. The comparison between the high- and low-immune groups showed that a total of 1,350 DEGs, including 1,290 upregulated genes and 60 downregulated genes, were identified (|log2 FC|>1, FDR <0.05) (Figure 2B). Similarly, in the stromal groups, 1,566 genes were upregulated and 157genes were downregulated in the high-score group (|log2 FC|>1, FDR <0.05) (Figure 2C). Distinct gene expression profiles were shown in heatmaps in the high- and low-score immune and stromal subgroups (Figure 2D,E). The Venn diagrams indicated 1,059 commonly upregulated genes in the high-score groups, and 33 commonly downregulated genes in the low-score groups (Figure 2F,G).
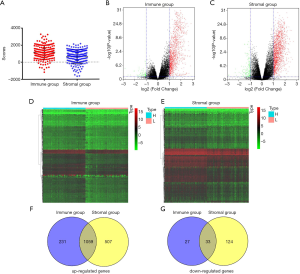
In the present study, considering that upregulated genes accounted for most of the DEGs, we focused on the collection of 1,797 upregulated genes and selected those for subsequent analysis. Of these, 1,566 genes were in the stromal group, while 1,290 genes were in the immune group. The upregulated genes here specifically referred to the DEGs in high-risk groups compared with low-risk groups, not tumor tissues versus normal tissues.
Functional enrichment analysis of DEGs
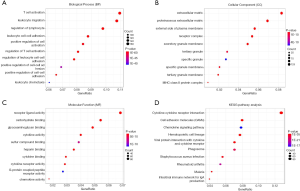
To explore the potential biologic function of the DEGs, functional enrichment analysis was performed for 1,797 genes. The top GO terms identified included T-cell activation, leukocyte migration, extracellular matrix, and receptor ligand activity (Figure 3A,B,C). KEGG analyses indicated that the DEGs were mainly enriched in the cytokine–cytokine receptor interaction, cell adhesion molecules (CAMs), and hematopoietic cell lineage pathway, demonstrating the reliability of the ESTIMATE algorithm (Figure 3D).
Construction and validation of prognostic risk score model for pancreatic cancer
To explore the role of DEGs in overall survival, we first performed a univariate Cox regression analysis for the selected genes and obtained a total of 279 prognosis-related genes, which reached statistical significance (P<0.05). The prognosis-related genes were further validated in an independent cohort, GSE62452, and 25 significantly and DEGs were identified (P<0.05). Subsequently, stepwise multivariate Cox regression analysis with 25 confirmed genes was performed using TCGA database; 7 genes were finally selected, and each regression coefficient was identified.
To effectively evaluate the prognosis of patients with pancreatic cancer, we constructed a prognostic risk model with the following formula:
Risk score =–1.1445 × expression of the polypeptide N-acetylgalactosaminyltransferase16 (GALNT16) - 0.5144 × expression of the corticotropin releasing hormone binding protein (CRHBP) + 0.7636 × expression of the C-X-C motif chemokine ligand 5 (CXCL5) –2.1059 × expression of the nucleotide binding oligomerization domain containing 2 (NOD2) +2.8607 × expression of the carbohydrate sulfotransferase 11 (CHST11) +1.0661 × expression of the zinc finger protein 683 (ZNF683) +0.4442 × expression of the mucin 16 (MUC16).
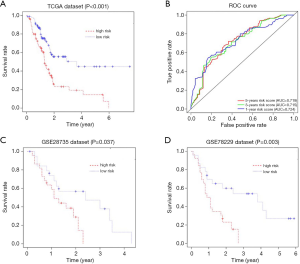
We then calculated the risk score for each patient. Patients were divided into 2 groups, a high-risk group and a low-risk group, based on the median value of the risk score according to the previous formula. Kaplan-Meier curves were used to evaluate the impact of risk score on the prognosis of pancreatic cancer patients from TCGA. Our results revealed that patients with a low-risk score had a longer survival time than patients with a high-risk score (P<0.001) (Figure 4A). The ROC curve analysis showed that the area under ROC curve (AUC) of our risk score model for 1-, 3- and 5-year-overall survival was 0.724, 0.715 and 0.719, respectively, suggesting a favorable efficiency in prognostic performance (Figure 4B). Remarkably, in accordance with the findings in TCGA, patients with high-risk scores had significantly worse OS than patients with low-risk score in the validation cohorts, GSE28735 and GSE78229, demonstrating the reliability and general applicability of our prognostic risk score model (P=0.037 and P=0.003, respectively (Figure 4C,D).
Comparison with conventional clinical characteristics
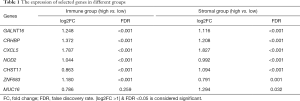
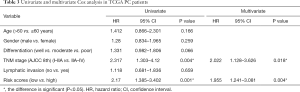
We first compared the expression of identified DEGs in different groups (immune and stromal groups). As shown in Table 1, GALNT16, CRHBP and CXCL5 were significantly upregulated in both the immune group and stromal groups. NOD2 and ZNF683 were found upregulated only in the immune group, while CHST11 and MUC16 upregulated in the stromal groups. Next, we compared the correlation between the risk score and conventional clinical factors. The results showed significantly correlation between our risk score and tumor differentiation (P<0.05) (Table 2). Subsequently we conducted univariate and multivariate COX regression analysis to explore the independent prognostic value of our risk score. Table 3 revealed that our risk score not only had a significant association with OS (P<0.05), but also remained an independent clinical predictor after adjusting for other clinical indicators.
Full table

Full table
Full table
Functional enrichment analysis and protein-protein interactions among genes of prognostic value
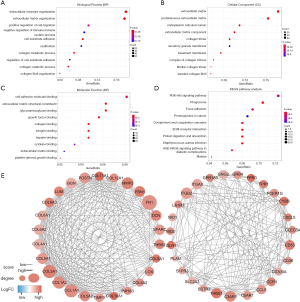
To better understand the functional implication of 7-genes prognostic model, a functional enrichment analysis was carried to illustrate their biological function. We identified 361 genes which were significantly correlated with 7 selected genes (Spearman |R| ≥0.6). GO analysis suggested that 7 selected genes and their co-expressed genes mainly focus on following terms: extracellular structure organization, extracellular matrix and cell adhesion molecule binding (Figure 5A,B,C). KEGG analysis showed that the top three signaling pathways were PI3K-Akt signaling pathway, Phagosome and Focal adhesion pathways (Figure 5D). Protein-protein interaction (PPI) networks were implemented to illustrate the interrelation among the identified DEGs. The network that consisted of 258 nodes and 1,291 interactions was made up of 11 modules. We selected the top two significant modules for further analysis (Figure 5E). In the first module which involved 26 nodes and 258 edges, FN1, COL1A1, COL3A1, COL1A2, COL5A1, BGN, and POSTN had the most connections with other members. Most of them are involved in extracellular matrix structural constituent. In the secondary module, ITGB2, C3AR1, THBS1, ITGAX, CD53, CCL5 had higher degree values, most of which are associated with immune responses.
Discussion
In the current work, we aim to identify prognosis-related genes on microenvironment in the TCGA database. By comparing the difference between immune score and stromal score basing on ESTIMATE algorithm, 1,797 up-regulated genes were extracted and confirmed to be involved in extracellular matrix and immune response by GO analysis and KEGG pathway analysis. Next, 7 genes were identified by univariate Cox and stepwise multivariate Cox regression analysis in TCGA and GSE62452 datasets and were selected to conduct a prognostic risk score model. Finally, the survival analysis in TCGA and GEO data set (GSE78229 and GSE28735) proved the reliability and wide availability of our prognostic risk model in predicting the prognosis.
The microenvironment of PDAC is distinguished from other malignancies by its abundance of extracellular matrix components and multiple types of innate and adaptive immune cells (23,24). Numerous activated carcinoma-associated fibroblasts, infiltrating immune cells, endothelial cells were found around tumour cells, forming a immunosuppressive tumor microenvironment with poor tissue perfusion. Moreover, in the occurrence and development of pancreatic cancer, the TME is constantly changing in composition and function, as to form a microenvironment conducive to proliferation and survival, metastasis, resistance to chemotherapy, affecting the prognosis of patients (25-28). Traditional treatments have produced limited benefits for prognosis of the PDCA patients so far. With the in-depth research on tumor microenvironment in recent years, immunotherapy, with its effective therapeutic response, controlled, and relative safety, has gradually become a hot spot in tumor studies. Currently, immunotherapy for pancreatic cancer mainly includes immune checkpoint inhibitors, cancer vaccines, adoptive cell transfer, combination with other immunotherapeutic agents (29). Many scholars have focused on the combinations of immunotherapy with traditional treatment, which is expected to be a new option for treatment methods.
During the past decades, many of pancreatic prognostic models about TME have been reported. For example, Moffitt et al. identified two stromal subtypes associated with different prognosis (30). Tsujikawa et al. described the prognostic value of immune cells via a multiplexed immunohistochemical platform (31). However, the complexity or limitations of those models limited their clinical application. Here, we tried to propose a feasible and operable TME-related prognostic model based on the RNA-Seq of tumor tissue by combining the two aspects of immune cells and stromal cells. Therefore, we explored the relationship between TME-related genes and prognosis of PADC patients. In our study, a prognostic model on TME was proposed and was verified effectively for pancreatic cancer.
Our prognostic risk score model was made up of seven TME-related genes. Among them, MUC16 (CA125) gene is a protein-coding gene expressing a heavily glycosylated type-I transmembrane mucin and overexpressed in multiple malignancies including pancreatic cancer (32). Muniyan et al. reported that MUC16 played an important role in pancreatic tumor development and could facilitate the metastasis of tumor cells (33). In our study, MUC16 was up-regulated in tumor tissues with high stromal scores. However, there was few reports on the role of MUC16 in the TME and further clinical research was need. ZNF683 (HOBIT) had demonstrated to be a core regulatory factor that controlled generation, proliferation, and survival of NK-cell progenitors and was crucial for generation of NK cell (34). Interestingly, our study found that, unlike what has been reported in other tumors (35), high expression of ZNF683 tended to be associated with poor prognosis in PDAC. CXCL5, as a chemokine that connects the extracellular microenvironment to the tumor (36), can be involved in the recruitment of immune cells by binding to their receptors (37) and contribute to the proliferation, migration and invasion of cancer cells (38-41). CRHBP is a corticotrophin releasing hormone binding protein gene. In the kidney cancer, CRHBP can promote the proliferation, invasion and metastasis of tumor cells by enhancing NF-κB and p53-mediated apoptosis induction (42). CHST11 was a key enzyme in the biosynthesis of chondroitin sulfates which was distributed on the surfaces of tumor cells and extracellular matrices (43,44). One study showed that CHST11 could play a role in the regulation of metastasis and chemoresistance via MAPK signal pathway (45). NOD2, a member of the intracellular NOD-like receptor family, had emerged as critical players in inducing proinflammatory and antimicrobial responses (46) and was assumed to be associated with the tumorigenesis of many kinds of cancer, such as pancreatic, gastric, colorectal, gallbladder, biliary tract and liver cancer (47). GALNT16 is an N-acetylgalactosaminyltransferase gene that plays a role in altering protein O-glycosylation, which had great importance in tumor development (48). In this experiment, GALNT16 was associated with both immunity and matrix. Considering that there were few studies on GALNT16, we need to conduct further experiments to verify its function on TME.
There is certain drawback to this study. First, it only uses computer to simulate data and lacks the validation by specific clinical data, so an independent cohort study is need. On the other hand, the exactly mechanism of the seven genes in the oncogenesis and development of PDAC, especially on TME, remains to be fully elucidated. Further research will be considered to explore these mechanisms. Nevertheless, the combined analysis of TCGA and GEO databases shows that our model has great advantages in accuracy and stability for predicting the prognosis in microenvironment. Our prognostic risk score model is expected to be applied to clinical prognosis assessment of PDAC patients.
Conclusions
Our seven-genes risk score model on TME is a promising biomarker which could be used to predict the prognosis in PDAC patients. However, further studies are required on the mechanism of the TME. Meanwhile, prospective studies are needed to validate the accuracy for estimating prognoses in PDAC patients.
Acknowledgments
Funding: This research was funded by grants from the National Natural Science Foundation of China, grant number 81702951, 81370059, 81672807.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/gs-21-28
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/gs-21-28). All authors report grants from National Natural Science Foundation of China, during the conduct of the study. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Availability of Data and Materials: GEO datasets: https://www.ncbi.nlm.nih.gov/gds/. TCGA dataset:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Wolfgang CL, Herman JM, Laheru DA, et al. Recent progress in pancreatic cancer. CA Cancer J Clin 2013;63:318-48. [Crossref] [PubMed]
- Conroy T, Hammel P, Hebbar M, et al. FOLFIRINOX or Gemcitabine as Adjuvant Therapy for Pancreatic Cancer. N Engl J Med 2018;379:2395-2406. [Crossref] [PubMed]
- Şenbabaoğlu Y, Gejman RS, Winer AG, et al. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome Biol 2016;17:231. [Crossref] [PubMed]
- Dougan SK. The Pancreatic Cancer Microenvironment. Cancer J. 2017;23:321-5. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000;100:57-70. [Crossref] [PubMed]
- Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 2012;21:309-22. [Crossref] [PubMed]
- Winslow S, Lindquist KE, Edsjo A, et al. The expression pattern of matrix-producing tumor stroma is of prognostic importance in breast cancer. BMC Cancer 2016;16:841. [Crossref] [PubMed]
- Curry JM, Sprandio J, Cognetti D, et al. Tumor microenvironment in head and neck squamous cell carcinoma. Semin Oncol 2014;41:217-34. [Crossref] [PubMed]
- Kenny PA, Lee GY, Bissell MJ. Targeting the tumor microenvironment. Front Biosci 2007;12:3468-74. [Crossref] [PubMed]
- Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. [Crossref] [PubMed]
- Zhou R, Zeng D, Zhang J, et al. A robust panel based on tumour microenvironment genes for prognostic prediction and tailoring therapies in stage I-III colon cancer. EBioMedicine 2019;42:420-30. [Crossref] [PubMed]
- Fu R, Han CF, Ni T, et al. A ZEB1/p53 signaling axis in stromal fibroblasts promotes mammary epithelial tumours. Nat Commun 2019;10:3210. [Crossref] [PubMed]
- Jia D, Li S, Li D, Xue H, Yang D, Liu Y. Mining TCGA database for genes of prognostic value in glioblastoma microenvironment. Aging (Albany NY) 2018;10:592-605. [Crossref] [PubMed]
- Chen H, Kong Y, Yao Q, et al. Three hypomethylated genes were associated with poor overall survival in pancreatic cancer patients. Aging (Albany NY) 2019;11:885-97. [Crossref] [PubMed]
- Liao X, Huang K, Huang R, et al. Genome-scale analysis to identify prognostic markers in patients with early-stage pancreatic ductal adenocarcinoma after pancreaticoduodenectomy. Onco Targets Ther 2017;10:4493-506. [Crossref] [PubMed]
- Ito K, Murphy D. Application of ggplot2 to Pharmacometric Graphics. CPT Pharmacometrics Syst Pharmacol 2013;2:e79 [Crossref] [PubMed]
- Metsalu T, Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res 2015;43:W566-70 [Crossref] [PubMed]
- Oliveros JC. An interactive tool for comparing lists with Venn's diagrams. Venny. 2007-2015.
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017;45:D362-8. [Crossref] [PubMed]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003;4:2. [Crossref] [PubMed]
- Tomita Y, Azuma K, Nonaka Y, et al. Pancreatic fatty degeneration and fibrosis as predisposing factors for the development of pancreatic ductal adenocarcinoma. Pancreas 2014;43:1032-41. [Crossref] [PubMed]
- Neesse A, Frese KK, Bapiro TE, et al. CTGF antagonism with mAb FG-3019 enhances chemotherapy response without increasing drug delivery in murine ductal pancreas cancer. Proc Natl Acad Sci U S A 2013;110:12325-30. [Crossref] [PubMed]
- Haqq J, Howells LM, Garcea G, et al. Pancreatic stellate cells and pancreas cancer: current perspectives and future strategies. Eur J Cancer 2014;50:2570-82. [Crossref] [PubMed]
- Erez N, Truitt M, Olson P, et al. Cancer-Associated Fibroblasts Are Activated in Incipient Neoplasia to Orchestrate Tumor-Promoting Inflammation in an NF-kappaB-Dependent Manner. Cancer Cell 2010;17:135-47. [Crossref] [PubMed]
- Whatcott C, Han H, Posner RG, et al. Tumor-stromal interactions in pancreatic cancer. Crit Rev Oncog 2013;18:135-51. [Crossref] [PubMed]
- Feig C, Gopinathan A, Neesse A, et al. The pancreas cancer microenvironment. Clin Cancer Res 2012;18:4266-76. [Crossref] [PubMed]
- Schizas D, Charalampakis N, Kole C, et al. Immunotherapy for pancreatic cancer: A 2020 update. Cancer Treat Rev 2020;86:102016 [Crossref] [PubMed]
- Moffitt RA, Marayati R, Flate EL, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet 2015;47:1168-78. [Crossref] [PubMed]
- Tsujikawa T, Kumar S, Borkar RN, et al. Quantitative Multiplex Immunohistochemistry Reveals Myeloid-Inflamed Tumor-Immune Complexity Associated with Poor Prognosis. Cell Rep 2017;19:203-17. [Crossref] [PubMed]
- Haridas D, Chakraborty S, Ponnusamy MP, et al. Pathobiological implications of MUC16 expression in pancreatic cancer. PLoS One 2011;6:e26839 [Crossref] [PubMed]
- Muniyan S, Haridas D, Chugh S, et al. MUC16 contributes to the metastasis of pancreatic ductal adenocarcinoma through focal adhesion mediated signaling mechanism. Genes Cancer 2016;7:110-24. [Crossref] [PubMed]
- Post M, Cuapio A, Osl M, et al. The Transcription Factor ZNF683/HOBIT Regulates Human NK-Cell Development. Front Immunol 2017;8:535. [Crossref] [PubMed]
- Mami-Chouaib F, Blanc C, Corgnac S, et al. Resident memory T cells, critical components in tumor immunology. J Immunother Cancer 2018;6:87. [Crossref] [PubMed]
- Itatani Y, Kawada K, Inamoto S, et al. The Role of Chemokines in Promoting Colorectal Cancer Invasion/Metastasis. Int J Mol Sci 2016;17:643. [Crossref] [PubMed]
- Zhang W, Wang H, Sun M, et al. CXCL5/CXCR2 axis in tumor microenvironment as potential diagnostic biomarker and therapeutic target. Cancer Commun (Lond) 2020;40:69-80. [Crossref] [PubMed]
- Zhao J, Ou B, Han D, et al. Tumor-derived CXCL5 promotes human colorectal cancer metastasis through activation of the ERK/Elk-1/Snail and AKT/GSK3beta/beta-catenin pathways. Mol Cancer 2017;16:70. [Crossref] [PubMed]
- Roca H, Jones JD, Purica MC, et al. Apoptosis-induced CXCL5 accelerates inflammation and growth of prostate tumor metastases in bone. J Clin Invest 2018;128:248-66. [Crossref] [PubMed]
- Zhou SL, Dai Z, Zhou ZJ, et al. CXCL5 contributes to tumor metastasis and recurrence of intrahepatic cholangiocarcinoma by recruiting infiltrative intratumoral neutrophils. Carcinogenesis 2014;35:597-605. [Crossref] [PubMed]
- Zhang H, Xia W, Lu X, et al. A novel statistical prognostic score model that includes serum CXCL5 levels and clinical classification predicts risk of disease progression and survival of nasopharyngeal carcinoma patients. PLoS One 2013;8:e57830 [Crossref] [PubMed]
- Yang K, Xiao Y, Xu T, et al. Integrative analysis reveals CRHBP inhibits renal cell carcinoma progression by regulating inflammation and apoptosis. Cancer Gene Ther 2020;27:607-18. [Crossref] [PubMed]
- Mikami T, Mizumoto S, Kago N, et al. Specificities of three distinct human chondroitin/dermatan N-acetylgalactosamine 4-O-sulfotransferases demonstrated using partially desulfated dermatan sulfate as an acceptor: implication of differential roles in dermatan sulfate biosynthesis. J Biol Chem 2003;278:36115-27. [Crossref] [PubMed]
- Uyama T, Ishida M, Izumikawa T, et al. Chondroitin 4-O-sulfotransferase-1 regulates E disaccharide expression of chondroitin sulfate required for herpes simplex virus infectivity. J Biol Chem 2006;281:38668-74. [Crossref] [PubMed]
- Zhou H, Li Y, Song X, et al. CHST11/13 Regulate the Metastasis and Chemosensitivity of Human Hepatocellular Carcinoma Cells Via Mitogen-Activated Protein Kinase Pathway. Dig Dis Sci 2016;61:1972-85. [Crossref] [PubMed]
- Negroni A, Pierdomenico M, Cucchiara S, et al. NOD2 and inflammation: current insights. J Inflamm Res 2018;11:49-60. [Crossref] [PubMed]
- Kutikhin AG. Role of NOD1/CARD4 and NOD2/CARD15 gene polymorphisms in cancer etiology. Hum Immunol 2011;72:955-68. [Crossref] [PubMed]
- Wu H, He G, Song T, et al. Evaluation of GALNT16 polymorphisms to breast cancer risk in Chinese population. Mol Genet Genomic Med 2019;7:e848 [Crossref] [PubMed]
(English Language Editor: R. Scott)



