
The expression and clinical significance of signal transducer and activator of transcription 3, tumor necrosis factor α induced protein 8-like 2, and runt-related transcription factor 1 in breast cancer patients
Introduction
Breast cancer is the most common of all female malignant tumors, and the incidence has been rising in recent years. At present, there are many treatments for breast cancer, including radiotherapy, chemotherapy, surgical resection, and combined treatment with Chinese and Western medicine, all of which can have certain therapeutic effects. However, due to the low rates of early breast cancer diagnosis, the prognosis of most patients is not ideal. Therefore, in-depth research on the mechanisms, diagnosis, and treatment of breast cancer is of great significance for improving the prognosis of patients and their overall survival.
Signal transducer and activator of transcription 3 (STAT3) is a bifunctional protein that exists in the cytoplasm and is coupled to the tyrosine phosphorylation signaling pathway which is involved in cell proliferation, apoptosis, and other processes (1). Research shows that STAT3 can be used as a potential therapeutic target for triple-negative breast cancer (2). miR-634 is able to regulate STAT3 and enhance the sensitivity of breast cancer cells to radiation (3). miR-124 reverses the doxorubicin resistance of breast cancer stem cells through STAT3/HIF-1 signaling pathways (4). Tumor necrosis factor α induced protein 8-like 2 (TIPE2) is a recently discovered inflammatory regulator (5,6) that has a low level of expression in patients with malignant tumors such as colon cancer and thyroid cancer. However, the relationship between TIPE2 and breast cancer is unclear. Tumor necrosis factor-α (TNF-α) induces TIPE2 inhibits proliferation and tumorigenesis in breast cancer cells (7). TIPE2 suppresses breast cancer tumorigenesis, growth and metastasis possibly via regulation of the AKT and p38 signaling pathways (8). TIPE2 inhibits breast cancer development and metastasis possibly via promoting CD8+ T and NK cell-mediated antitumor immune responses (9). Runt-related transcription factor 1, (RUNX1) is located on chromosome 21p22 and contains 9 exons. It has been reported to be closely related to the occurrence of malignant tumors (10).The novel interplay between YAP, RUNX1 and RUNX3 and its significance in breast cancer progression can serve as a prognostic tool to predict cancer recurrence (11). miR-378-mediated suppression of Runx1 alleviates the aggressive phenotype of triple-negative MDA-MB-231 human breast cancer cells (12). In this study, the expression levels, as well as the clinical significance of STAT3, TIPE2, and RUNX1 in breast cancer tissues, were explored. We present the following article in accordance with the STARD reporting checklist (available at http://dx.doi.org/10.21037/gs-21-108).
Methods
General information
Breast cancer patients who underwent radical mastectomy for breast cancer in our hospital from October 2014 to October 2017 were enrolled in this study. This study was approved by the Sichuan Provincial People’s Hospital (East Hospital), University of Electronic Science and Technology of China (No. 20180529). All patients agreed to participate in this study and signed an informed consent form. The research complies with the requirements of the Declaration of Helsinki (as revised in 2013). The following inclusion criteria were applied: (I) patients were diagnosed with breast cancer by pathological examination combined with in-hospital laboratory examination (13); (II) the clinical data and imaging data were complete; and (III) patients showed good treatment compliance and could cooperate with medical staff to complete the follow-up. The following patients were excluded from the study: (I) patients with other malignant tumors; (II) patients with cognitive impairment; and (III) patients who had been administered drugs that can affect the levels of STAT3, TIPE2, and RUNX1 prior to the start of the study. According to the inclusion and exclusion criteria, the study eventually included 68 patients (68 breast cancer tissue specimens) as the observation group, and the corresponding normal healthy tissue 3 cm away from the cancer tissue was selected as the control group.
Study method
Measurement of STAT3, TIPE2, RUNX1 levels
Immunohistochemical staining was used to determine the expression levels of STAT3, TIPE2, and RUNX1 following standard protocols. Specifically, specimens were embedded in paraffin blocks and 4 µm thick sections were baked in a constant temperature box at 75 °C for over 6 hours. Specimens were removed and allowed to cool naturally before dewaxing using xylene and an ethanol gradient (100%, 90%, and 75%). Hydrogen peroxide solution (3%) was then used to eliminate the endogenous peroxidase activity. After dewaxing, the specimens were placed in sodium citrate buffer for heat repair (15 minutes), and following natural cooling, a goat serum blocking solution was added dropwise in the wet box to prevent non-specific binding. After settling for 20 minutes, the rabbit anti-human TIPE2 polyclonal antibody (1:400), rabbit anti-human RUNX1 monoclonal antibody (1:400), and rabbit anti-human STAT3 monoclonal antibody (1:400) were added dropwise. A phosphate buffered saline (PBS) solution was used for the negative control. The specimens were then placed in a 4 °C incubator overnight. Thereafter, the specimens were re-warmed, washed with PBS, and a biotin-labeled goat anti-rabbit IgG and horseradish enzyme-labeled streptavidin were added dropwise. After incubating for 20 minutes at room temperature, a diaminobenzidine color solution was added. Samples were then rinsed with tap water and counterstained with hematoxylin, before dehydration and sealing with neutral resin. The rabbit anti-human TIPE2 polyclonal antibody, rabbit anti-human STAT3 monoclonal antibody, and rabbit anti-human RUNX1 monoclonal antibody were all purchased from Abcam, UK.
Western blot analysis
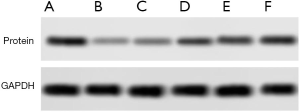
The proteinwas extracted respectively. After quantitative analysis by BCA method, the sample buffer was added. After protein boiling, the sample was loaded and electrophoresis was carried out. The protein expression levels of STAT3, TIPE2, RUNX1 were detected, and the gray value of the item was analyzed by image studio software. The ratio of the gray value of the target protein band to the gray value of the internal reference band was used as the expression amount of the target protein, each group repeated three times, and the follow-up statistical analysis was carried out.
Judgment criteria
A total of 5 fields of view were randomly chosen and observed by 3 pathologists under an optical microscope. Each field contained at least 100 intact tumor cells. The determination criteria for positive STAT3, TIPE2, RUNX1 staining (14) were based on the proportion of positive cells in the tissue cells and the degree of staining. Specifically, the scoring system for the proportion of positive cells was as follows: 0 points for ≤5%, 1 point for >5–25%, 2 points for >25–50%, 3 points for >50–75%, and 4 points for >75%. The scoring system for the staining intensity was as follows: 0 points for undyed, 1 point for light yellow, 2 points for brown, and 3 points for tan. Staining index = proportion of positive cells score + staining intensity score. A staining index >3 was classified as high expression and a staining index ≤3 was classified as low expression.
Follow-up results
The follow-up period of this study was 2 years. Follow-up commenced after the patient underwent a radical mastectomy and ended with the conclusion of this study (October 2019) or death of the patient. The follow-up time ranged from 7 to 24 months, with an average follow-up time of 15.11±2.87 months. Follow-up methods included a visit to the hospital for review or telephone consultation.
Statistical analysis
SPSS22.0 software was used for statistical analysis. Measurement data were presented as mean ± standard deviation (
Results
A comparison of STAT3, TIPE2, and RUNX1 expression levels in breast cancer tissue and healthy tissue
Expression of STAT3 was significantly higher in the observation group compared to the control group. In contrast, the expression of TIPE2 and RUNX1 was significantly lower in the observation group compared to control group (P<0.05) as shown in Table 1 and Figures 1,2.

Full table

Univariate analysis of the 2-year survival of breast cancer patients
Of the 68 patients, 51 survived within the 2-year follow-up period. However, 17 patients died. Univariate analysis demonstrated that age, tumor diameter, and histological type did not affect the 2-year prognosis and survival of breast cancer patients (P>0.05). In contrast, positive STAT3 expression, negative TIPE2 expression, negative RUNX1 expression, poor histological differentiation, TNM stage III–IV, and distant metastasis were all factors that affected the 2-year prognosis and survival of breast cancer patients (P<0.05) as shown in Table 2.

Full table
Multivariate analysis of the 2-year prognosis and survival of breast cancer patients
Multivariate analyses revealed that positive STAT3 expression, negative TIPE2 expression, negative RUNX1 expression, poor histological differentiation, TNM stage III–IV, and distant metastasis were all independent risk factors that can affect the 2-year prognosis of breast cancer patients (P<0.05; Table 3).

Full table
The predictive value of STAT3, TIPE2, and RUNX1 status, and the combined value of the three factors in the 2-year prognostic survival of breast cancer patients
The ROC curve was used to analyze the predictive value of STAT3, TIPE2, and RUNX1 levels, and the combined detection of the three factors on the 2-year prognostic survival of breast cancer patients. The area under the curve of STAT3, TIPE2, and RUNX1 expression, as well as the combination of the 3 factors were 0.806, 0.760, 0.688, and 0.910, respectively. The area under the curve achieved the largest value when all 3 proteins were combined as showed in Table 4 and Figure 3.

Full table

Discussion
The biogenesis and development of breast cancer are associated with changes in multiple genes, including the activation of oncogenes and the loss of tumor suppressor gene function, which result in unregulated cell proliferation, differentiation, and inhibition of apoptosis (15-18). Current clinical efforts are devoted to identifying highly-specific breast cancer-related genes. The combined detection of multiple breast cancer-related genes may lead to improved rates of early breast cancer diagnosis, targeted therapy, and prognostic risk assessment, all of which have important clinical significance for the prognosis of patients with breast cancer.
STAT3 is an important gene involved in the self-renewal of human embryonic stem cells, and it is mostly expressed in epithelial malignant tumors (19). Studies have shown that STAT3 can promote tumor cell proliferation and metastasis, and inhibit anti-tumor immune responses (20), all of which are closely related to the prognosis of tumor patients. The results of this current study demonstrated that the expression of STAT3 and TIPE2 in breast cancer tissues are quite different. The positive rate of STAT3 expression was significantly higher than that of adjacent normal tissues, while the positive rate of TIPE2 expression was significantly lower than that of adjacent normal tissues. This is consistent with previous reports (21) and suggested that these two proteins may be important indicators for the early detection of breast cancer. Studies have also shown that upregulation of STAT3 expression can promote the proliferation of cancer stem cells and inhibit their differentiation (22,23). IL-6/STAT3 signaling hijacks estrogen receptor α enhancers to drive breast cancer metastasis (24). Inhibition of STAT3 signaling pathway by natural product pectolinarigenin attenuates breast cancer metastasis (25). Not surprisingly, the results of our study also revealed that higher rates of positive STAT3 expression were associated with poorer prognosis of the patient. Therefore, these results suggested that inhibiting the expression of STAT3 may effectively prevent the proliferation and differentiation of tumor cells, thereby improving the prognosis of patients.
In recent years, the relationship between TIPE2 protein expression and early malignant tumor cell proliferation has gradually attracted attention. Some studies reported that in the process of early malignant tumor cell proliferation, TIPE2 plays a role in reducing inflammation (26). Tumor necrosis factor-α (TNF-α) induces TIPE2 inhibits proliferation and tumorigenesis in breast cancer cells (7). TIPE2 suppresses breast cancer tumorigenesis, growth and metastasis possibly via regulation of the AKT and p38 signaling pathways (8).TIPE2 inhibits breast cancer development and metastasis possibly via promoting CD8+ T and NK cell-mediated antitumor immune responses (9). This current study demonstrated that low expression of TIPE2 is an independent risk factor for the 2-year prognosis of survival in breast cancer patients. The decrease in TIPE2 protein expression can be related to the decreased infiltration ability of cancer cells, the decreased ability to break through the basement membrane, or the decreased ability of lymph node adhesion. The absence or decrease of TIPE2 expression in breast cancer tissue may play an important role in breast cancer development.
RUNX1 can either activate or inhibit related genes or signaling pathways in different solid malignant tumors, and thus it can play either a carcinogenic or tumor suppressor role. Indeed, RUNX1 has been shown to be carcinogenesis or tumor suppressive in different hematological tumors (27). Rada et al. (28) reported that RUNX1 had a high rate of positive expression in epithelial tumors such as ovarian cancer, and head and neck squamous cell carcinomas, and thus it functions as an oncogene. In contrast, Bustos et al. (29) demonstrated that RUNX1 was expressed at low levels in breast and colorectal cancer tissues, and thus may act as a tumor suppressor gene. By comparing the expression of RUNX1 in breast cancer tissues and the adjacent normal tissues, our study found that the expression of RUNX1 in breast cancer tissues was significantly lower than that in adjacent normal tissues, further confirming the tumor suppressor role of RUNX1 in breast cancer. In addition, patients with lower expression of RUNX1 had a significantly higher prognostic probability of death at 2 years after surgery compared to patients with higher expression of RUNX1. This suggested that RUNX1 has a significant role in the prognosis of breast cancer patients. The mechanisms of RUNX1 in apoptosis and tumorigenesis are complicated, and are yet to be fully elucidated (30).
RUNX1 may repress ts-112 to prevent overactive proliferation in breast epithelial cells to augment its established roles in maintaining the mammary epithelium (31). Future follow-up studies should conduct in-depth research on the specific molecular mechanisms of RUNX1 in the apoptosis and drug resistance of breast cancer cells. This will provide novel insights for the clinical implementation of targeted therapy for breast cancer patients. The ROC curve analysis examining the 2-year prognostic survival of breast cancer patients showed that the area under the curve achieved the largest value when the predictive values of STAT3, TIPE2, and RUNX1 were combined. This suggested that the combined detection of these three genes may be an effective prognostic marker for breast cancer patients following surgery.
In summary, the expression levels of STAT3, TIPE2, RUNX1 in breast cancer tissues is significantly different from that in adjacent normal tissues, and the combined detection of STAT3, TIPE2, and RUNX1 may improve the rate of early breast cancer diagnosis and prognosis. This in turn will facilitate timely and specific treatments so as to improve the overall survival of breast cancer patients.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at http://dx.doi.org/10.21037/gs-21-108
Data Sharing Statement: Available at http://dx.doi.org/10.21037/gs-21-108
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/gs-21-108). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Sichuan Provincial People’s Hospital (East Hospital), University of Electronic Science and Technology of China (No. 20180529). All patients agreed to participate in this study and signed an informed consent form. The research complies with the requirements of the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Greer S, Morris T, Pettingale KW. Psychological response to breast cancer: effect on outcome. Lancet 1979;2:785-7. [Crossref] [PubMed]
- Qin JJ, Yan L, Zhang J, et al. STAT3 as a potential therapeutic target in triple negative breast cancer: a systematic review. J Exp Clin Cancer Res 2019;38:195. [Crossref] [PubMed]
- Yang B, Kuai F, Chen Z, et al. miR-634 Decreases the Radioresistance of Human Breast Cancer Cells by Targeting STAT3. Cancer Biother Radiopharm 2020;35:241-8. [Crossref] [PubMed]
- Liu C, Xing H, Guo C, et al. MiR-124 reversed the doxorubicin resistance of breast cancer stem cells through STAT3/HIF-1 signaling pathways. Cell Cycle 2019;18:2215-27. [Crossref] [PubMed]
- Marmot MG, Altman DG, Cameron DA, et al. The benefits and harms of breast cancer screening: an independent review. Br J Cancer 2013;108:2205-40. [Crossref] [PubMed]
- Sovak MA, Bellas RE, Kim DW, et al. Aberrant nuclear factor-kappaB/Rel expression and the pathogenesis of breast cancer. J Clin Invest 1997;100:2952-60. [Crossref] [PubMed]
- Wang K, Ren Y, Liu Y, et al. Tumor Necrosis Factor (TNF)-α-Induced Protein 8-like-2 (TIPE2) Inhibits Proliferation and Tumorigenesis in Breast Cancer Cells. Oncol Res 2017;25:55-63. [Crossref] [PubMed]
- Zhang Z, Liu L, Liu C, et al. TIPE2 suppresses the tumorigenesis, growth and metastasis of breast cancer via inhibition of the AKT and p38 signaling pathways. Oncol Rep 2016;36:3311-6. [Crossref] [PubMed]
- Zhang Z, Liu L, Cao S, et al. Gene delivery of TIPE2 inhibits breast cancer development and metastasis via CD8(+) T and NK cell-mediated antitumor responses. Mol Immunol 2017;85:230-7. [Crossref] [PubMed]
- Yasuda MT, Sakakibara H, Shimoi K. Estrogen- and stress-induced DNA damage in breast cancer and chemoprevention with dietary flavonoid. Genes Environ 2017;39:10. [Crossref] [PubMed]
- Kulkarni M, Tan TZ, Syed Sulaiman NB, et al. RUNX1 and RUNX3 protect against YAP-mediated EMT, stem-ness and shorter survival outcomes in breast cancer. Oncotarget 2018;9:14175-92. [Crossref] [PubMed]
- Browne G, Dragon JA, Hong D, et al. MicroRNA-378-mediated suppression of Runx1 alleviates the aggressive phenotype of triple-negative MDA-MB-231 human breast cancer cells. Tumour Biol 2016;37:8825-39. [Crossref] [PubMed]
- Martellosio A, Pasian M, Bozzi M, et al. Dielectric Properties Characterization From 0.5 to 50 GHz of Breast Cancer Tissues. IEEE Transactions on Microwave Theory & Techniques 2017;65:998-1011. [Crossref]
- Kang MH, Jeong KJ, Kim WY, et al. Musashi RNA-binding protein 2 regulates estrogen receptor 1 function in breast cancer. Oncogene 2017;36:1745-52. [Crossref] [PubMed]
- Borstelmann NA, Rosenberg S, Gelber S, et al. Partners of young breast cancer survivors: a cross-sectional evaluation of psychosocial concerns, coping, and mental health. J Psychosoc Oncol 2020;38:670-86. [Crossref] [PubMed]
- Stein MJ, Karir A, Arnaout A, et al. Quality-of-Life and Surgical Outcomes for Breast Cancer Patients Treated with Therapeutic Reduction Mammoplasty Versus Mastectomy with Immediate Reconstruction. Ann Surg Oncol 2020;27:4502-12. [Crossref] [PubMed]
- Skrobo D, Walsh N, Quinn C, et al. 173P Clinical characteristics of long-term responders to anti-HER2 therapy in metastatic breast cancer: A review of the charactHER clinical data. Ann Oncol 2020;31:S78. [Crossref]
- Kilcar AY, Yildiz O, Dogan T, et al. The Effect of Bitter Melon (Momordica charantia) Extract on the Uptake of 99mTc Labeled Paclitaxel: In Vitro Monitoring in Breast Cancer Cells. Anticancer Agents Med Chem 2020;20:1497-503. [Crossref] [PubMed]
- Sorace AG, Syed AK, Barnes SL, et al. Quantitative [18F]FMISO PET Imaging Shows Reduction of Hypoxia Following Trastuzumab in a Murine Model of HER2+ Breast Cancer. Mol Imaging Biol 2017;19:130-7. [Crossref] [PubMed]
- McDonald PJ, Carpenter R, Royle GT, et al. Poor response of breast cancer to tamoxifen. Postgrad Med J 1990;66:1029-31. [Crossref] [PubMed]
- Chen H, Yong-Jie J, Heng-Xiao W. Signal transduction and transcriptional activator 3 involved in pathogenesis and development of cervical cancer. Chinese Journal of Immunology 2019;20:78-85.
- Ridgley LA, Anderson AE, Maney NJ, et al. IL-6 Mediated Transcriptional Programming of Naïve CD4+ T Cells in Early Rheumatoid Arthritis Drives Dysregulated Effector Function. Front Immunol 2019;10:1535. [Crossref] [PubMed]
- Mitrofan CG, Appleby SL, Nash GB, et al. Bone morphogenetic protein 9 (BMP9) and BMP10 enhance tumor necrosis factor-α-induced monocyte recruitment to the vascular endothelium mainly via activin receptor-like kinase 2. J Biol Chem 2017;292:13714-26. [Crossref] [PubMed]
- Siersbæk R, Scabia V, Nagarajan S, et al. IL6/STAT3 Signaling Hijacks Estrogen Receptor α Enhancers to Drive Breast Cancer Metastasis. Cancer Cell 2020;38:412-23.e9. [Crossref] [PubMed]
- Li Y, Gan C, Zhang Y, et al. Inhibition of Stat3 Signaling Pathway by Natural Product Pectolinarigenin Attenuates Breast Cancer Metastasis. Front Pharmacol 2019;10:1195. [Crossref] [PubMed]
- Hofmann D, Salmon L, Wider G. Activity of Tumor Necrosis Factor α Is Modulated by Dynamic Conformational Rearrangements. J Am Chem Soc 2018;140:167-75. [Crossref] [PubMed]
- Pontejo SM, Sanchez C, Ruiz-Argüello B, Alcami A. Insights into ligand binding by a viral tumor necrosis factor (TNF) decoy receptor yield a selective soluble human type 2 TNF receptor. J Biol Chem 2019;294:5214-27. [Crossref] [PubMed]
- Rada M, Kapelanski-Lamoureux A, Petrillo S, et al. Runt related transcription factor-1 (runx1) plays a central role in vessel co-option of colorectal cancer liver metastases. Cancer Res 2020; [Crossref]
- Bustos F, Sepúlveda H, Prieto CP, et al. Runt-Related Transcription Factor 2 Induction During Differentiation of Wharton's Jelly Mesenchymal Stem Cells to Osteoblasts Is Regulated by Jumonji AT-Rich Interactive Domain 1B Histone Demethylase. Stem Cells 2017;35:2430-41. [Crossref] [PubMed]
- Kaptan E, Sancar Bas S, Sancakli A, et al. Runt-Related Transcription Factor 2 (Runx2) Is Responsible for Galectin-3 Overexpression in Human Thyroid Carcinoma. J Cell Biochem 2017;118:3911-9. [Crossref] [PubMed]
- Farina NH, Scalia S, Adams CE, et al. Identification of tRNA-derived small RNA (tsRNA) responsive to the tumor suppressor, RUNX1, in breast cancer. J Cell Physiol 2020;235:5318-27. [Crossref] [PubMed]
(English Language Editor: J. Teoh)


