
Identification of novel hub genes and lncRNAs related to the prognosis and progression of pancreatic cancer by microarray and integrated bioinformatics analysis
Introduction
Pancreatic cancer (PC) is one of the most aggressive human digestive cancers and has an extremely poor prognosis, demonstrating a 5-year survival rate of only 10% (1). Despite this, there has been little progress in the diagnosis and treatment of PC in the past two decades. Due to the lack of early symptoms and diagnosis, approximately 80% of PC patients lose the opportunity to have radical surgery because of the primary tumor’s invasion of the adjacent organs or surrounding vital vessels, and many patients exhibit extensive metastasis even at their first diagnosis (2). In spite of continuing research, the mechanisms of PC development remain unclear. Therefore, there is an urgent need to explore the regulating mechanisms in PC proliferation, invasion, and metastasis, and to identify novel effective prognostic and therapeutic targets for PC.
Emerging evidence has confirmed that, in regard to hub genes, many long non-coding RNAs (lncRNAs) may also play core roles in PC (3-5) and other malignant phenotypes of multiple tumors and could represent novel targets for diagnosis and treatment (6-8).
In this study, we used an mRNA-lncRNA microarray expression analysis to determine differentially expressed genes (DEGs) and differentially expressed lncRNAs (DElncRNAs) in PC. To detect hub genes and hub lncRNAs in PC, we utilized a bioinformatics approach to analyze those DEGs and DElncRNAs. Using the Enrichr database, Gene Ontology (GO) analysis was conducted for the functional annotation assessment, and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was conducted for the enrichment assessment. LncRNA-mRNA co-expressed networks were constructed to explore the core regulating factors. The GEPIA and ONCOMINE databases were then used to identify hub genes which were associated with both overall survival and tumor stage in PC. Ultimately, we found five hub genes and sixteen hub lncRNAs that were strongly linked to the development and progression of PC. In this study, we used commercial microarrays and integrated bioinformatics analysis to screen out hub DEGs and DElncRNAs, which most of them never been reported before in PC. Different from co-expressed networks conducted by online databases, we established lncRNA-mRNA co-expressed networks in PC and paired adjacent non-tumor samples also considering their differential expressions, resulting a more convinced result. Furthermore, via these bioinformatics analysis, we map hub DElncRNAs as potential ceRNAs of hub genes, which can indicating our next experimental direction.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/gs-21-151).
Methods
Tissue samples
A total of five pairs of PC and paired adjacent non-tumor (AN) samples were obtained from PC patients who received surgery between 2012 and 2013 in the Department of Pancreatic Surgery at Changhai Hospital, Second Military Medical University, Shanghai, China. Among the PC patients, three were male and two were female, aging from 46 to 72. None of the patients in this study had received any neoadjuvant anti-tumor therapies. Two certified pathologists confirmed all diagnoses, which were based on pathological evidence. All samples were frozen immediately in liquid nitrogen with a TRIzol reagent (Invitrogen) after the tumor was removed surgically and were then transferred to a −80 °C refrigerator until required.
This study was approved by the Ethics Committee for Biomedical Research of the Second Military Medical University. Informed consent was obtained from all patients or their relatives. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
LncRNA-mRNA microarrays
The frozen PC samples were delivered to KangChen Biotechnology Co. Ltd (Shanghai, China) for the mRNAs and lncRNAs extraction, quantity testing, labeling and analysis. In order to profile both human mRNAs and lncRNAs, the Arraystar Human mRNA-LncRNA Array v2.0 was designed. Array images were screened and analyzed using the Agilent Feature Extraction software (version 10.7.3.1), and acquired data was normalized and processed by the GeneSpring GX v11.5.1 software package (Agilent Technologies). LncRNAs and mRNAs, which were flagged as present or marginal in no less than half of the samples, were identified for further analysis. DElncRNAs and DEGs between the PC tissues and matched AN samples were confirmed through fold change filtering. Paired t-tests were performed to compare the mean differences. P-value/FDR and Volcano Plot filtering were used to identify the statistical significance of the DElncRNAs and DEGs between the PC tissues and matched AN samples. Hierarchical clustering analysis was conducted by the Cluster 3.0 and JAVA Treeview software packages. P<0.05 and |log2FC| >1 were considered statistically significant.
Enrichr database analysis
The Enrichr database (http://amp.pharm.mssm.edu/Enrichr) was used to perform the GO functional annotation and KEGG pathways enrichment analysis of the DEGs (9,10). Biological process (BP), cellular component (CC), and molecular function (MF) were included in the GO functional annotation. The top ten enriched GO functional annotations and KEGG pathways of DEGs were displayed as images downloaded from web pages. P<0.05 was considered statistically significant.
LncRNA-mRNA co-expressed network establishment
LncRNA-mRNA co-expressed networks in both PC and AN tissues were constructed to detect the correlations between DEGs and DElncRNAs (11) according to their normalized signal intensity of specific expression. The Pearson correlation was calculated for each pair of mRNA-lncRNA, mRNA-mRNA, and lncRNA-lncRNA (12). As the simplest and most important measure of an mRNA or lncRNA, indicating the centrality and relative importance in a network, degree centrality is defined as the number of associations one profile has with the others (13). Core regulatory DEGs and DElncRNAs are considered to possess the greatest degree differences between the PC and AN networks. Results were ranked by the absolute relative degree value |DiffK| (relative degree value, PC-AN, DiffK).
ONCOMINE database analysis
The expressions of potential core genes were assessed in the PC tissues compared to those in normal pancreatic tissues in the ONCOMINE (https://www.oncomine.org/) datasets. ONCOMINE is the world’s largest microarray database and integrated data-mining platform of neoplasms at present. P<0.05 was considered statistically significant.
GEPIA database analysis
The Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/about.html) database is a newly developed interactive web server for analyzing RNA sequencing expression data of tumors and normal tissues obtained from The Cancer Genome Atlas (TCGA) and Genoytype-Tissue Expression (GTEx) databases. GEPIA now contains a total of 9,736 tumors and 8,587 normal samples (14). The GEPIA database was used to validate the expression levels of significant DEGs in PC. Cut-off criteria were determined as P<0.05 and |log2FC| >1. GEPIA was also used to identify the relationship between DEGs expressing levels and PC patients’ pathological stages and overall survival. P<0.05 was considered statistically significant.
Statistical analysis
Statistical analysis was done based on the online databases respectively. In lncRNA-mRNA microarrays, paired t-tests were performed to compare the mean differences. P-value/FDR and Volcano Plot filtering were used to identify the statistical significance of the DElncRNAs and DEGs between the PC tissues and matched AN samples. P<0.05 and |log2FC| >1 were considered statistically significant.
Results
Identification of DEGs and lncRNAs
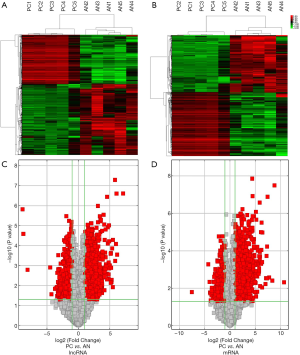
To explore the DEGs and DElncRNAs in PC, we had previously collected five pairs of PC samples from patients who had undergone surgery, and we used microarrays to analyze the DEGs and DElncRNAs between the PC tissues and paired AN tissues. (GEO, http://www.ncbi.nlm.nih.gov/geo/, ID: GSE101094). There were 943 mRNAs differentially expressed, of which 501 were up-regulated and 442 were down-regulated (fold change >2, P<0.05, https://cdn.amegroups.cn/static/public/10.21037gs-21-151-1.xlsx). We confirmed that 1,138 lncRNA transcripts were differentially expressed, comprising 470 up-regulated and 668 down-regulated lncRNAs (fold change >2, P<0.05, https://cdn.amegroups.cn/static/public/10.21037gs-21-151-2.xlsx).
Hierarchical clustering analysis showed that the DEGs and DElncRNAs between the PC tissues and paired AN tissues were systematically variated (Figure 1A,B). In addition, volcano plots were constructed to identify the DEG and DElncRNA transcripts (Figure 1C,D). The results suggested that the DEGs and DElncRNAs were likely representative and most probably played a role in regulating the development of PC.
GO annotation and KEGG pathway enrichment analyses
To determine the potential biological roles of the above 943 DEGs, we used the online Enrichr database to conduct the GO annotation and KEGG pathway enrichment analyses. The annotated results for the GO analyses contained three parts: BP, MF, and CC. GO BP analysis revealed that the top three significantly enriched terms of the up-regulated DEGs were adrenal gland development (GO:0030325), epithelial cell migration (GO:0010631), and peptide hormone secretion (GO:0030072); the top three enriched terms of the down-regulated DEGs were positive regulation of MAPK cascade (GO:0043410), organic anion transport (GO:0015711), and regulation of calcium ion transport (GO:0051924). In the GO MF analysis, the 501 up-regulated DEGs were mainly enriched in nuclear hormone receptor binding (GO:0035257), transmembrane receptor protein serine/threonine kinase binding (GO:0070696), and cadherin binding (GO:0045296); the 442 down-regulated DEGs were mainly enriched in sodium-independent organic anion transmembrane transporter activity (GO:0015347), organic anion transmembrane transporter activity (GO:0008514), and G-protein coupled photoreceptor activity (GO:0008020). In the GO CC analysis, the up-regulated DEGs were mostly enriched in focal adhesion (GO:0005925), the cytoplasmic vesicle membrane (GO:0030659), and the endoplasmic reticulum lumen (GO:0005788); the down-regulated DEGs were mostly enriched in the integral component of the plasma membrane (GO:0005887), the endocytic vesicle (GO:0030139), and the spanning component of the plasma membrane (GO:0044214) (Figure 2A,B,C,D,E,F).
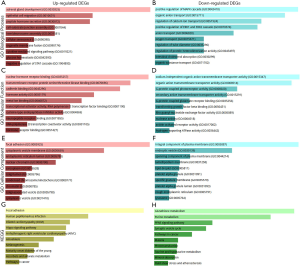
The top three markedly enriched KEGG pathways for the 501 up-regulated DEGs were focal adhesion, human papillomavirus infection, and dilated cardiomyopathy (DCM); the 442 down-regulated DEGs were mostly enriched in glutathione metabolism, purine metabolism, and the PPAR signaling pathways (Figure 2G,H).
LncRNA-mRNA co-expressed network establishment and hub gene identification
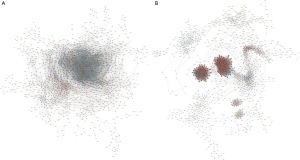
In order to locate the core regulatory factors, we constructed lncRNA-mRNA co-expressed networks of PC tissues and AN tissues (Figure 3A,B, https://cdn.amegroups.cn/static/public/10.21037gs-21-151-3.xlsx, https://cdn.amegroups.cn/static/public/10.21037gs-21-151-4.xlsx). In a co-expressed network, core functional transcripts mostly have more relationships with other factors than nonfunctional transcripts, and always have the largest absolute relative degree values. Thus, mRNAs with the top 100 largest absolute values of relative degree differences between the PC and AN lncRNA-mRNA co-expressed networks (https://cdn.amegroups.cn/static/public/10.21037gs-21-151-5.xlsx) were selected as the potential hub genes for further study.
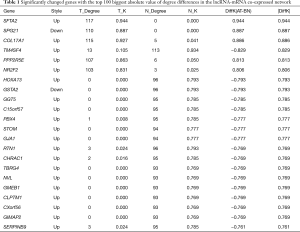
We compared these transcriptional levels of genes in PC tissues and normal pancreatic tissues by using the ONCOMINE and GEPIA databases, and identified that 22 mRNA levels, among the top 100 genes confirmed above, were significantly differentially expressed between PC and normal tissues, with the same expressed trend in our microarrays. They were SFTA2, SPG21, COL17A1, TM4SF4, PPP2R5E, NR2F2, HOXA13, GSTA2, GGT5, C15orf57, PBX4, STOM, GJA1, RTN1, CHRAC1, TBRG4, NVL, GMEB1, CLPTM1, CXorf56, GIMAP2, and SERPINB9 (Tables 1,2, Figure 4). Among them, HOXA13 and NVL were only identified as differentially expressed in the ONCOMINE database, and PPP2R5E, GSTA2, C15orf57, PBX4, CHRAC1, GMEB1, CLPTM1, and CXorf56 were only confirmed to be differentially regulated in the GEPIA database.
Full table
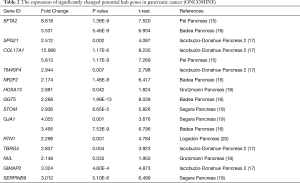
Full table

The relationship of hub genes with overall survival and tumor stage
We next used the GEPIA database to study the association between overall survival and the 22 genes identified above by the median and quartile of their expression (Figure S1). We noticed that high levels of SFTA2 (median: P=0.0089, HR =1.7; Quartile: P=0.00042, HR =3), SPG21 (median: P=0.015, HR =1.7; quartile: P=0.00042, HR =1.7), COL17A1 (median: P=0.00039, HR =2.1; quartile: P=0.00052, HR =2.8), PPP2R5E (quartile: P=0.027, HR =1.9), HOXA13 (median: P=0.007, HR =1.8), and CHRAC1 (quartile: P=0.027, HR =1.9) were associated with worse overall survival in PC. Conversely, high levels of RTN1 (median: P=0.0039, HR =0.55; quartile: P=0.0024, HR =0.41) and CLPTM1 (median: P=0.0081, HR =0.57; quartile: P=0.013, HR =0.47) were linked with improved overall survival in PC.
The GEPIA pathological stage plot analysis revealed that the levels of SFTA2, COL17A1, HOXA13, PBX4, RTN1, CHRAC1, GMEB1, and CXorf56 varied significantly according to the tumor stage of the PC (Figure S1).
As a result of these findings, we considered SFTA2, COL17A1, HOXA13, RTN1, and CHRAC1 to be the hub genes in PC, as their expression levels were associated with both overall survival and tumor stage.
Identification of hub lncRNAs
In order to identify the potential core lncRNAs in PC, we examined the co-expression correlations between DElncRNAs and DEGs in both the PC and AN tissues. We identified the lncRNAs with the most significant correlations to the five hub genes identified above (Pearson correlation coefficient >0.999, https://cdn.amegroups.cn/static/public/10.21037gs-21-151-6.xlsx). Of these potential core lncRNAs, we then selected 16 lncRNAs which were also within the top 100 largest absolute value of degree differences in the lncRNA-mRNA co-expressed network (https://cdn.amegroups.cn/static/public/10.21037gs-21-151-7.xlsx). As a result, we identified AL109748.6, AC125792.1, CTD-2292M14.1, AK027298, AK094441, BC084573, RP11-288L9.1, AC012005.5, NR_002827, NR_024058, NR_024427, AK056486, RP11-312H15.3, AC006427.3, BC035067, and CTD-2066L21.3 as the hub lncRNAs in PC (Table 3).
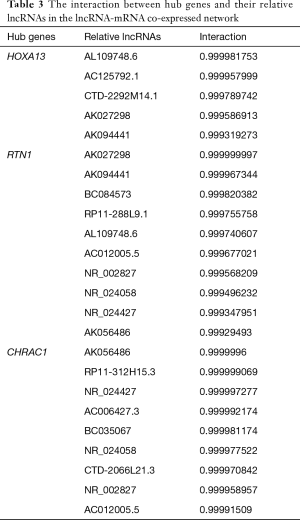
Full table
Discussion
PC is one of the most life-threatening diseases in the world. In recent decades significant advances have been made in understanding the molecular mechanisms of PC and in improving treatment, but to date no sensitive biomarker has been identified that could assist with early diagnosis or provide an effective target for drug therapy, and the mortality rate remains close to 100% (21). Therefore, the identification of new factors underlying PC development and novel individualized therapeutic targets are urgently needed for PC patients.
In the present study, we used commercial microarrays to screen out DEGs and DElncRNAs between human PC and paired AN tissues. Subsequent bioinformatics analyses were performed to identify the hub genes and hub lncRNAs correlated with PC. As a result, 943 DEGs and 1,138 DElncRNAs were identified. KEGG enrichment analysis revealed that focal adhesion was the most significantly enriched pathway for 501 up-regulated DEGs, and glutathione metabolism was the most significantly enriched pathway for 442 down-regulated DEGs. In cell biology, focal adhesions create combinatorial signaling complexes and mediate the actin-integrin function to regulate cellular behaviors such as cell motility, proliferation, differentiation, survival, and regulation of gene expression (22,23). Thus, increased activation of focal adhesions is closely associated with tumor metastasis and clinical prognosis, especially in PC. Shi et al. found that loss of linc01060 could induce PC invasion and metastasis through a vinculin-mediated focal adhesion turnover (24). Ennajdaoui et al. identified that IGF2BP3 could alter the focal adhesion junction and promote PC cell invasiveness (25). Focal adhesion kinase (FAK) is known to be a scaffolding protein and an integral component of focal adhesions. Furuyama et al. suggested that FAK was upregulated in PC and was correlated with the size of PC (26). Moreover, glutathione is a vital intracellular antioxidant. Enzymes of glutathione metabolism have been found to be mediated by oncogenic Kras in PC resulting in a dysregulation of PC growth both in vitro and in vivo (27). Chio et al. found that AKT signaling inhibition and glutathione synthesis could prolong the survival of PC cells, suggesting a prospective target for therapy (28). As a result, it would appear that both focal adhesion and glutathione metabolism are essential pathways in PC progression, as we have identified with DEGs in PC via microarray and KEGG pathway enrichment analyses.
Based on the lncRNA-mRNA co-expressed network, we confirmed SFTA2, COL17A1, HOXA13, RTN1, and CHRAC1 as the hub genes in PC, and their expression levels were associated with both overall survival and tumor stage, verified by the ONCOMINE and GEPIA databases. Previous studies have reported that SFTA2 (surfactant-associated 2), which is a member of the surfactant protein family, assembles innate immune responses in the central nervous system and lung tissues (29,30). It was also identified as a core regulating gene related to carcinogenesis and prognosis in colorectal cancer (31,32). However, its detailed mechanism is still unclear. COL17A1 (collagen type XVII alpha 1 chain), a collagen family member, maintains an integral part of the hemidesmosome structure. It was established as a novel gene upregulated in a dog bladder cancer organoid culture (33) and has been shown to be increased in multiple neoplasms, such as ovarian cancer, breast cancer, and lung cancer (34,35). COL17A1 was also identified as a p53 downstreaming transcriptional target in breast tissues that inhibits cell migration and invasion and is associated with better prognosis (36). Moreover, this gene was linked to overall survival and classic AJCC staging of PC patients in TCGA and the STRING online tool and external validation datasets (37,38). HOXA13 is a member of the homeobox A cluster (HOXA) gene family with prognostic significance and is involved in a wide range of biological functions in human cancers such as esophageal squamous cell carcinoma, laryngeal squamous cell cancer, ovarian cancer, prostate cancer, and gastric cancer (39-43). It has also been shown to promote PC progression and gemcitabine resistance via a HOTTIP-HOXA13 axis route (44). RTN1 (reticulon 1) belongs to the reticulon-encoding gene family and is considered as a specific marker and potential therapeutic target for non-small-cell lung cancers, colorectal cancer, and prostate cancer (45-48). CHRAC1 (Chromatin Accessibility Complex Subunit 1) is a histone-fold protein-encoding gene, functioning in DNA transcription, replication, and packaging as a sequence-specific DNA binding component, and associated with survival progression among breast cancer patients (49,50). However, little is known of the mechanisms of these five novel hub genes in PC, and, to the best of our knowledge, SFTA2, RTN1, and CHRAC1 have not previously been reported in PC.
In order to comprehensively analyze the functions of DElncRNAs in the development and progression of PC and their status in the PC-regulating network, we mapped DElncRNAs in the lncRNA-mRNA co-expressed network of PC and AN tissues. A variety of interactions among DEGs and DElncRNAs were obtained. We identified hub lncRNAs with the largest absolute value of degree differences in a co-expressed network, and with the most significant correlation with the five novel hub genes identified above. As a result, we confirmed 16 hub lncRNAs in PC, which were AL109748.6, AC125792.1, CTD-2292M14.1, AK027298, AK094441, BC084573, RP11-288L9.1, AC012005.5, NR_002827, NR_024058, NR_024427, AK056486, RP11-312H15.3, AC006427.3, BC035067, and CTD-2066L21.3. Few functions and mechanisms of these hub lncRNAs have been known to us in human diseases. Among them, AK056486 has been reported down-regulated in lesional skin biopsies compared to non-lesional ones (51). RP11-288L9.4 can regulate HCV antibiral innate immunity via JAK-STAT signaling pathway (52). However, RP11-288L9.1 is still unclear currently.
As lncRNAs have been reported to work as ceRNAs in regulating the expression and function of genes, 16 hub lncRNAs were also indicated as ceRNAs with their correlated hub genes. As most of these hub lncRNAs have not been reported in PC before, additional experimental and clinical validation is required to support our findings.
Currently, the targeted treatment for pancreatic cancer and the study of its microenvironment is still at the early stage. As in in the newest NCCN clinical practice guideline of pancreatic adenocarcinoma, olaparib has been recommended for patients with germline BRCA1/2 mutations, pembrolizumab has been recommended for MSI-H or dMMR tumors, larotrectinib and Entrectinib could been used if NTRK gene fusion positive in pancreatic cancer patients during metastatic stage as the first-line therapy (53). However, the positive rate of sequencing is quite low as far as our clinical experience in Chinses patients. On the other aspect, the effects of immunotherapies, such as CTLA-4 monoclonal antibody, CAR-T therapy and tumor vaccine, are still unsatisfactory in pancreatic cancer (54-56). As a result, further studies for the core factors in pancreatic cancer development and progression are urgently needed to develop newly therapies and diagnostic targets.
In summary, using an mRNA-lncRNA microarray assay of PC and paired AN tissues and integrated bioinformatics analysis, we identified DEGs and DElncRNAs in PC and constructed mRNA-lncRNA co-expressed regulatory networks. We further indicated five novel hub genes and sixteen novel related hub lncRNAs with potentially significant predictive and therapeutic values in PC. Further clinical studies with larger sample sizes, together with experimental studies focused on the detailed molecular mechanisms involved, are needed to validate the present findings.
Acknowledgments
Funding: This work was supported by the grant from Shanghai Changzheng Hospital Foundation (YQ676).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/gs-21-151
Data Sharing Statement: Available at http://dx.doi.org/10.21037/gs-21-151
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/gs-21-151). All authors report that this work was supported by the grant from Shanghai Changzheng Hospital Foundation (YQ676); the fee of microarray was supported by grants from the Science and Technology Commission of Shanghai Municipality (124119a0501); and payment for manuscript writing is provided by Shanghai Changzheng Hospital. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee for Biomedical Research of the Second Military Medical University. Informed consent was obtained from all patients or their relatives. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Grossberg AJ, Chu LC, Deig CR, et al. Multidisciplinary standards of care and recent progress in pancreatic ductal adenocarcinoma. CA Cancer J Clin 2020;70:375-403. [Crossref] [PubMed]
- Poruk KE, Firpo MA, Adler DG, et al. Screening for pancreatic cancer: why, how, and who? Ann Surg 2013;257:17-26. [Crossref] [PubMed]
- Liang X, Qi M, Wu R, et al. Long non-coding RNA CUDR promotes malignant phenotypes in pancreatic ductal adenocarcinoma via activating AKT and ERK signaling pathways. Int J Oncol 2018;53:2671-82. [Crossref] [PubMed]
- Lei S, He Z, Chen T, et al. Long noncoding RNA 00976 promotes pancreatic cancer progression through OTUD7B by sponging miR-137 involving EGFR/MAPK pathway: Journal of experimental & clinical cancer research. CR 2019;38:470. [PubMed]
- Peng WX, He RZ, Zhang Z, et al. LINC00346 promotes pancreatic cancer progression through the CTCF-mediated Myc transcription. Oncogene 2019;38:6770-80. [Crossref] [PubMed]
- Xiong XD, Ren X, Cai MY, et al. Long non-coding RNAs: An emerging powerhouse in the battle between life and death of tumor cells. Drug Resist. Updat. 2016;26:28-42. [Crossref] [PubMed]
- Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol 2013;26:155-65. [Crossref] [PubMed]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nature reviews. Genetics 2009;10:155-9. [Crossref] [PubMed]
- Chen EY, Tan CM, Kou Y, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 2013;14:128. [Crossref] [PubMed]
- Kuleshov MV, Jones MR, Rouillard AD, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016;44:W90-7. [Crossref] [PubMed]
- Pujana MA, Han JD, Starita LM, et al. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat Genet 2007;39:1338-49. [Crossref] [PubMed]
- Prieto C, Risueno A, Fontanillo C, et al. Human gene coexpression landscape: confident network derived from tissue transcriptomic profiles. PLoS One 2008;3:e3911 [Crossref] [PubMed]
- Barabási AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat Rev Genet 2004;5:101-13. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Li L, Zhang JW, Jenkins G, et al. Genetic variations associated with gemcitabine treatment outcome in pancreatic cancer. Pharmacogenet Genomics 2016;26:527-37. [Crossref] [PubMed]
- Badea L, Herlea V, Dima SO, et al. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology 2008;55:2016-27. [PubMed]
- Iacobuzio-Donahue CA, Maitra A, Olsen M, et al. Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. Am J Pathol 2003;162:1151-62. [Crossref] [PubMed]
- Grützmann R, Pilarsky C, Ammerpohl O, et al. Gene expression profiling of microdissected pancreatic ductal carcinomas using high-density DNA microarrays. Neoplasia 2004;6:611-22. [Crossref] [PubMed]
- Segara D, Biankin AV, Kench JG, et al. Expression of HOXB2, a retinoic acid signaling target in pancreatic cancer and pancreatic intraepithelial neoplasia. Clin Cancer Res 2005;11:3587-96. [Crossref] [PubMed]
- Logsdon CD, Simeone DM, Binkley C, et al. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res 2003;63:2649-57. [PubMed]
- Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med 2014;371:1039-49. [Crossref] [PubMed]
- Wozniak MA, Modzelewska K, Kwong L, et al. Focal adhesion regulation of cell behavior. Biochim Biophys Acta 2004;1692:103-19. [Crossref] [PubMed]
- Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nature reviews. Nat Rev Mol Cell Biol 2009;10:21-33. [Crossref] [PubMed]
- Shi X, Guo X, Li X, et al. Loss of Linc01060 induces pancreatic cancer progression through vinculin-mediated focal adhesion turnover. Cancer Lett 2018;433:76-85. [Crossref] [PubMed]
- Ennajdaoui H, Howard JM, Sterne-Weiler T, et al. IGF2BP3 Modulates the Interaction of Invasion-Associated Transcripts with RISC. Cell Rep 2016;15:1876-83. [Crossref] [PubMed]
- Furuyama K, Doi R, Mori T, et al. Clinical significance of focal adhesion kinase in resectable pancreatic cancer. World J Surg 2006;30:219-26. [Crossref] [PubMed]
- Son J, Lyssiotis CA, Ying H, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 2013;496:101-5. [Crossref] [PubMed]
- Chio IIC, Jafarnejad SM, Ponz-Sarvise M, et al. NRF2 Promotes Tumor Maintenance by Modulating mRNA Translation in Pancreatic Cancer. Cell 2016;166:963-76. [Crossref] [PubMed]
- Krause M, Peukert N, Härtig W, et al. Localization, Occurrence, and CSF Changes of SP-G, a New Surface Active Protein with Assumable Immunoregulatory Functions in the CNS. Mol Neurobiol 2019;56:2433-9. [Crossref] [PubMed]
- Xiao J, Lu X, Chen X, et al. Eight potential biomarkers for distinguishing between lung adenocarcinoma and squamous cell carcinoma. Oncotarget 2017;8:71759-71. [Crossref] [PubMed]
- Li C, Shen Z, Zhou Y, et al. Independent prognostic genes and mechanism investigation for colon cancer. Biol Res 2018;51:10. [Crossref] [PubMed]
- Gong B, Kao Y, Zhang C, et al. Identification of Hub Genes Related to Carcinogenesis and Prognosis in Colorectal Cancer Based on Integrated Bioinformatics. Mediators Inflamm 2020;2020:5934821 [Crossref] [PubMed]
- Elbadawy M, Usui T, Mori T, et al. Establishment of a novel experimental model for muscle-invasive bladder cancer using a dog bladder cancer organoid culture. Cancer Sci 2019;110:2806-21. [Crossref] [PubMed]
- López-Ayllón BD, de Castro-Carpeño J, Rodriguez C, et al. Biomarkers of erlotinib response in non-small cell lung cancer tumors that do not harbor the more common epidermal growth factor receptor mutations. Int J Clin Exp Pathol 2015;8:2888-98. [PubMed]
- Januchowski R, Świerczewska M, Sterzyńska K, et al. Increased Expression of Several Collagen Genes is Associated with Drug Resistance in Ovarian Cancer Cell Lines. J Cancer 2016;7:1295-310. [Crossref] [PubMed]
- Yodsurang V, Tanikawa C, Miyamoto T, et al. Identification of a novel p53 target, COL17A1, that inhibits breast cancer cell migration and invasion. Oncotarget 2017;8:55790-803. [Crossref] [PubMed]
- Wu M, Li X, Zhang T, et al. Identification of a Nine-Gene Signature and Establishment of a Prognostic Nomogram Predicting Overall Survival of Pancreatic Cancer. Front Oncol 2019;9:996. [Crossref] [PubMed]
- Wu J, Li Z, Zeng K, et al. Key genes associated with pancreatic cancer and their association with outcomes: A bioinformatics analysis. Mol Med Rep 2019;20:1343-52. [Crossref] [PubMed]
- Li J, Ye M, Zhou C. Expression Profile and Prognostic Values of Family Members in Laryngeal Squamous Cell Cancer. Front Oncol 2020;10:368. [Crossref] [PubMed]
- Kelly Z, Moller-Levet C, McGrath S, et al. The prognostic significance of specific HOX gene expression patterns in ovarian cancer. Int J Cancer 2016;139:1608-17. [Crossref] [PubMed]
- Luo Z, Rhie SK, Lay FD, et al. A Prostate Cancer Risk Element Functions as a Repressive Loop that Regulates HOXA13. Cell Rep 2017;21:1411-7. [Crossref] [PubMed]
- Gu ZD, Shen LY, Wang H, et al. HOXA13 promotes cancer cell growth and predicts poor survival of patients with esophageal squamous cell carcinoma. Cancer Res 2009;69:4969-73. [Crossref] [PubMed]
- Wu DC, Wang SSW, Liu C-J, et al. Reprogramming Antagonizes the Oncogenicity of HOXA13-Long Noncoding RNA HOTTIP Axis in Gastric Cancer Cells. Stem Cells 2017;35:2115-28. [Crossref] [PubMed]
- Li Z, Zhao X, Zhou Y, et al. The long non-coding RNA HOTTIP promotes progression and gemcitabine resistance by regulating HOXA13 in pancreatic cancer. J Transl Med 2015;13:84. [Crossref] [PubMed]
- Woenckhaus M, Klein-Hitpass L, Grepmeier U, et al. Smoking and cancer-related gene expression in bronchial epithelium and non-small-cell lung cancers. J Pathol 2006;210:192-204. [Crossref] [PubMed]
- van de Velde HJ, Senden NH, Roskams TA, et al. NSP-encoded reticulons are neuroendocrine markers of a novel category in human lung cancer diagnosis. Cancer Res 1994;54:4769-76. [PubMed]
- Lemire M, Qu C, Loo LWM, et al. A genome-wide association study for colorectal cancer identifies a risk locus in 14q23.1. Hum Genet 2015;134:1249-62. [Crossref] [PubMed]
- Hamzeh O, Alkhateeb A, Zheng J, et al. Prediction of tumor location in prostate cancer tissue using a machine learning system on gene expression data. BMC Bioinformatics 2020;21:78. [Crossref] [PubMed]
- Mahmood SF, Gruel N, Chapeaublanc E, et al. A siRNA screen identifies RAD21, EIF3H, CHRAC1 and TANC2 as driver genes within the 8q23, 8q24.3 and 17q23 amplicons in breast cancer with effects on cell growth, survival and transformation. Carcinogenesis 2014;35:670-82. [Crossref] [PubMed]
- Lin W, Lin HD, Guo XY, et al. Allelic expression imbalance polymorphisms in susceptibility chromosome regions and the risk and survival of breast cancer. Mol Carcinog 2017;56:300-11. [Crossref] [PubMed]
- Zouboulis CC, Nogueira da Costa A, Makrantonaki E, et al. Alterations in innate immunity and epithelial cell differentiation are the molecular pillars of hidradenitis suppurativa. J Eur Acad Dermatol Venereol 2020;34:846-61. [Crossref] [PubMed]
- Liu X, Duan X, Holmes JA, et al. A Long Noncoding RNA Regulates Hepatitis C Virus Infection Through Interferon Alpha-Inducible Protein 6. Hepatology 2019;69:1004-19. [Crossref] [PubMed]
- Tempero MA, Malafa MP, Al-Hawary M, et al. Pancreatic Adenocarcinoma, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2021;
- Winograd R, Byrne KT, Evans RA, et al. Induction of T-cell Immunity Overcomes Complete Resistance to PD-1 and CTLA-4 Blockade and Improves Survival in Pancreatic Carcinoma. Cancer Immunol Res 2015;3:399-411. [Crossref] [PubMed]
- Le DT, Picozzi VJ, Ko AH, et al. Results from a Phase IIb, Randomized, Multicenter Study of GVAX Pancreas and CRS-207 Compared with Chemotherapy in Adults with Previously Treated Metastatic Pancreatic Adenocarcinoma (ECLIPSE Study). Clin Cancer Res 2019;25:5493-502. [Crossref] [PubMed]
- Stromnes IM, Schmitt TM, Hulbert A, et al. T Cells Engineered against a Native Antigen Can Surmount Immunologic and Physical Barriers to Treat Pancreatic Ductal Adenocarcinoma. Cancer Cell 2015;28:638-52. [Crossref] [PubMed]
(English Language Editor: D. Fitzgerald)


