
Identification of hub genes in triple-negative breast cancer by integrated bioinformatics analysis
Introduction
Breast cancer (BC) is a frequently diagnosed malignancy and a serious threat to women’s health (1). Although large numbers of breast cancer patients have benefited from advancements in medical treatments, high rates of recurrence and metastasis are the leading cause of death from cancer among women (2,3). There are multiple subtypes of BC, according to estrogen receptors, progesterone receptors, and human epidermal growth factor receptor 2 (Her2). The characteristic of triple negative breast cancer (TNBC) is deficiency in the estrogen receptors, progesterone receptors, and Her2 protein (4). In recent years, many attempts have been made to explore the biological therapy, targets of related molecular activity, and relevant signals for TNBC (5). Comprehensive study on the mechanism of TNBC is warranted for the advancement of its treatment and management. There is an increasing body of research on differential analysis of BC gene expression data to identify potential biomarkers of BC. Based on this, R software (https://r-project.org) and its related package were used to compare the gene expression profiles of TNBC and normal tissues, and to explore the potential molecular targets and related signaling pathways for the occurrence and development of TNBC at the genomic level. GEO is a public functional genomics database maintained by the NCBI for genomic analysis of a variety of diseases. Kaplan-Meier Plotter database is an online analysis database related to the prognosis of malignant tumors, which can be used to evaluate the effect of gene expression on the prognosis of cancer and to evaluate the relationship between target gene expression and related tumor prognosis. This provided an important theoretical basis for further discovery of new TNBC treatments. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/gs-21-17).
Methods
Data sources and processing and Gene Expression Omnibus database
To performed the gene expression matrix, Rstudio (version 3.6.3) supported by the R software platform (version 3.6.3) and relevant packages were applied. The microarray data of GSE76250, downloaded from the Gene Expression Omnibus (GEO) data set, was made up of gene expression data based on the Affymetrix Human Transcriptome Array 2.0 (Thermo Fisher Scientific, Waltham, MA, USA) gene expression array from 198 cases, involving 165 TNBC samples and 33 paired normal breast tissues. Firstly, microarray data were standardized, and then the probe with the highest average expression level was used to match with the corresponding genes by R function. These transformed genes representing the expression profiles were analyzed in this study. The study was conducted in accordance to with the Declaration of Helsinki (as revised in 2013).
Identification of differentially expressed genes (DEGs)
The limma package (version 3.42.2) was used to identify the differentially expressed genes (DEGs) betwixt TNBC tissues and normal samples. In this work, genes with a false discovery rate (FDR) <0.01 and log fold change (logFC) ≥1 or ≤−1 were considered DEGs. The ggplot2 package (version 1.26.0), a drawing tool in R software, was visualized to create a volcano plot and heatmap of all DEGs.
Enrichment analysis of DEGs
The enrichment analysis of DEGs was conducted through Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. The GO term and KEGG pathway enrichment analysis, based on the clusterProfiler package (version 3.14.3) in R software, contained biological process (BP), cellular component (CC), molecular function (MF), and KEGG pathway. Enrichment analysis results were visualized using the ggplot2 package (version 1.26.0). A P value <0.05 was considered statistically significant.
Protein-protein interaction network (PPIN) building and interrelation analysis of DEGs
The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (an online biological protein database, https://string-db.org/) was used for the analysis and establishment of PPINs and the localization of hub genes. Usually, hub genes are the most important in the whole network, and are highly connected with their modules and highly correlated to other genes in the same modules. In this work, all DEGs were uploaded to the STRING database to obtain the quantitative relationship between genes, and then PPIN was constructed using Cytoscape software [version 3.7.2, (https://cytoscape.org)]. Based on Cytoscape, MCODE was conducted to analyze modules from the PPIN. Modules with an MCODE score >60 were further analyzed, and the hub DEGs with a degree >80 were identified. Survival analysis of hub DEGs was based on Kaplan Meier-plotter (www.kmplot.com).
Statistical methods
A P value <0.05 was considered statistically significant. The FDR referred to the adjusted P-value, and FDR <0.01 was regarded as the threshold for screening DEGs. Hypergeometric testing and the Kaplan-Meier statistical method were used for enrichment analysis and survival analysis, respectively.
Results
Data pre-processing
The gene expression profile of GSE76250 from the GEO database was processed by the R software function, and the profile consisted of 165 TNBC samples and 33 paired normal breast tissue samples, including 70,523 microarray data. The microarray data were further standardized and subjected to genome-name conversion.
Identification of DEGs
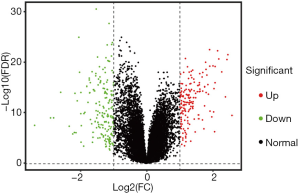
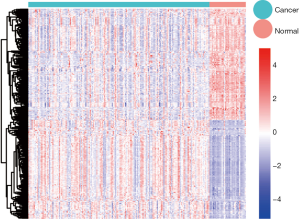
Based on the limma package of R language, the gene expression levels of TNBC and normal tissues were compared and analyzed to further identify the DEGs. A total of 340 DEGs were identified, among which 160 were up-regulated and 180 were down-regulated genes. The DEGs were selected based on the following criteria: FDR <0.01 and logFC ≥1 or logFC ≤−1. All 30,905 genes were visualized and are shown in a volcano plot (Figure 1), with red dots representing the 160 up-regulated genes and green dots representing the 180 down-regulated genes. As shown in Figure 2, the heatmap showed all significantly DEGs and their levels of expression.
Enrichment analysis of DEGs
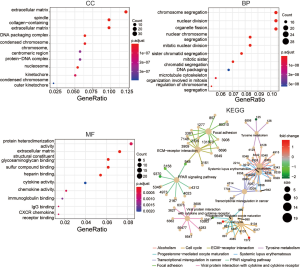
The GO and KEGG pathway analysis of DEGs were enforced by clusterProfiler package. The GO enrichment analyses were conducted at three aspects: CC, BP, and MF. As a result of GO and KEGG enrichment analysis, the DEGs were primarily located in extracellular matrix (ECM), spindle, collagen-containing ECM, chromosome segregation, nuclear division, organelle fission, protein heterodimerization activity, ECM structural constituent, glycosaminoglycan binding, cell cycle, ECM-receptor interaction, focal adhesion, respectively in CC, BP, MF, and KEGG groups, as shown in Figure 3.
Construction and analysis of PPIN for DEGs
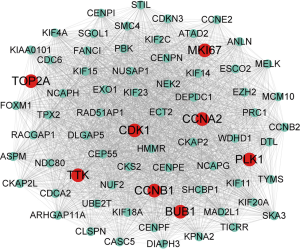
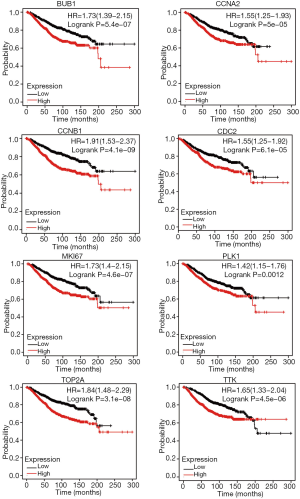
In this work, all DEGs were uploaded to the STRING database, and PPIN was set up. Then, the PPIN was visualized using Cytoscape software. A functional module that MCODE Score >64 and degree >80 was revealed, as shown in Figure 4, identifying 8 hub genes significantly related to other genes, including CCNB1, CDK1, TOP2A, MKI67, TTK, CCNA2, BUB1, and PLK1. Survival analysis showed that high gene expression indicted poor overall survival (OS), as shown in Figure 5.
Discussion
Characteristically, TNBC is aggressive and associated with a poor prognosis. The treatment options for patients with TNBC are limited, and currently only surgery and chemotherapy (alone or combined) are commonly available (6). Since not all cancer-driving genes have yet been identified, it is of great significance to explore the molecular mechanism of the occurrence and development of TNBC. Better biomarkers are urgently needed to predict cancer prognosis and progression. In the present study, DEGs obtained by comparing TNBC and normal tissues underwent further enrichment analysis, PPIN, and survival analysis, to explore potential biomarkers that affect the progression and prognosis of TNBC.
The function of enrichment analysis indicated that DEGs were related to the metastasis and occurrence of cancer cells. Cancer cells can reform or penetrate into the ECM composed the scaffold of tissues and organs, and the ECM can release growth factors and chemokines, eventually leading to distant metastasis of cancer cells (7). Tumor-derived ECM is biochemically distinct in its composition and is stiffer compared to the normal ECM; therefore, the composition of ECM could affect the treatment of tumors (8). In most cases, sudden fluctuations of human chromosomes content will have adverse consequences, such as meiotic aneuploidy, resulting in abnormal embryos or cancer cells developing into malignant tumors (9).
Hub genes were involved in the construction of spindle assembly checkpoints protein families, which mainly affect cell mitosis to maintain genome stability. Numerous studies have indicated that overexpression of BUB1 affects the progression and recurrence of many cancers, such as breast, familial colorectal, and hepatocellular carcinoma (10). The BUB1 encoded a threonine-protein kinase BubR1 is involved in the construction of spindle assembly checkpoints and was overexpressed in TNBC (11). These studies have confirmed that knocking out or silencing BUB1 inhibits the potential of TNBC stem cells, thereby inhibiting the formation of xenografts in immunocompromised mice (12-14). Cell death can also be mediated by BUB1 in response to chromosome mis-segregation, and BUB1 acts to suppress spontaneous tumorigenesis (15). Monopolar spindle 1 kinase (Mps1), also known as TTK protein kinase, is the core component of the spindle assembly checkpoint and plays a key role in regulating the process of cell division (16). With the exception of testes and placenta, it is difficult to detect transcripts of the TTK gene in normal organs. However, high levels of Mps1 can be found in many human malignancies including glioblastoma, thyroid cancer, and breast cancer (17).
Hub genes were also shown to mediate cell cycle regulation. The gene CCNB1, also named cycliB1, is part of a highly reactionary protein family of cyclins and plays significant roles in the progression and development of cancers (18-20). The pathways associated with CCNB1 include DNA double-strand repair and DNA damage response (21). It has been confirmed to be abnormally expressed in dozens of cancer types based on The Human Protein Atlas, indicating the feasible function of CCNB1 in cancer progression (22). It has been recorded that the high expression of CCNB1 in estrogen receptor positive breast cancer was significantly associated with poor hormone therapy outcomes (19). Trastuzumab emtansine (T-DM1) has induced defective CCNB1, resulting in acquired resistance, which serves as a pharmacodynamic predictor after T-DM1 treatment in Her2 positive breast cancer (23). The gene CCNA2 was identified as associated with the E2F transcription factor and has been reported to be a target for oncogenic signals (24,25). The protein encoded by CDK1 is a member of the serine (Ser) or threonine (Thr) protein kinase family and its key biological function is to control the eukaryotic cell cycle by regulating the centrosome cycle (26). Therefore, it is an effective therapeutic target for inhibitors in cancer therapy (27-29). At present, the treatment of BC requires precise and multi-targeted therapy (30). The birth of CDK4/6 inhibitors has benefited an increasing number of breast cancer patients. The birth of CDK1 may further improve the quality of life of cancer patients, and clinical trials have reported the evaluation of targeting CDK1 (31).
Hub genes were closely related to breast cancer cell proliferation and metastasis. The protein Ki67 is considered to play a key role in cell proliferation, and is expressed in the cell nucleus during the G1, S, G2, and M phase of the cell cycle, but not in the cell quiescent state (32). The expression level of Ki67 was detected by immunohistochemistry (IHC), which is called Ki67 index to assess cancer tissue proliferation (33). At present, Ki67 assessment is mainly used to estimate the prognosis of ER+/HER2 breast cancer, guide the choice of adjuvant therapy, and predict the efficacy of neoadjuvant therapy (34). In ER-/HER2+ and ER-/HER2- tumors, high Ki67 index after neoadjuvant therapy is associated with poor prognosis (34). The TOP2A gene encodes a DNA topoisomerase 2-alpha enzyme, whose main function is instrumental in DNA replication and transcription, control, and change of DNA topological state (35). It is seen as a subtle and particular biomarker of actively proliferating cells, and is closely associated with the proliferation and invasion of various types of tumors (36).
Conclusions
In general, these DEGs were shown to play important roles in a variety of BPs. The biological mechanisms of some hub genes in particular have been elucidated, some of which were shown to be concerned with the development and treatment of cancer.
In summary, this study involved a comprehensive bioinformatics analysis by comparing TNBC and paired normal gene expression profiles. The analysis resulted in the identification of the hub genes and pathways associated with TNBC. Furthermore, the hub genes were identified as highly expressed in BC tissues and were confirmed to be linked with the progression and poorer prognosis of TNBC. Our research results provided valuable references for basic and clinical studies of TNBC in the future. Further investigation of these genes is required.
Acknowledgments
The authors would like to extend their thanks to TCGA and GEO database creators and participants for providing open access to gene expression and clinical phenotype data to researchers. Thanks is also given to the authors who provided technical help for the research analysis.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/gs-21-17
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/gs-21-17). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance to with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Harbeck N, Penault-Llorca F, Cortes J, et al. Breast cancer. Nat Rev Dis Primers 2019;5:66. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Fan W, Chang J, Fu P. Endocrine therapy resistance in breast cancer: current status, possible mechanisms and overcoming strategies. Future Med Chem 2015;7:1511-9. [Crossref] [PubMed]
- Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med 2010;363:1938-48. [Crossref] [PubMed]
- Jhan JR, Andrechek ER. Triple-negative breast cancer and the potential for targeted therapy. Pharmacogenomics 2017;18:1595-609. [Crossref] [PubMed]
- Wahba HA, El-Hadaad HA. Current approaches in treatment of triple-negative breast cancer. Cancer Biol Med 2015;12:106-16. [PubMed]
- Eble JA, Niland S. The extracellular matrix in tumor progression and metastasis. Clin Exp Metastasis 2019;36:171-98. [Crossref] [PubMed]
- Nallanthighal S, Heiserman JP, Cheon DJ. The Role of the Extracellular Matrix in Cancer Stemness. Front Cell Dev Biol 2019;7:86. [Crossref] [PubMed]
- Potapova T, Gorbsky GJ. The Consequences of Chromosome Segregation Errors in Mitosis and Meiosis. Biology (Basel) 2017;6:12. [Crossref] [PubMed]
- Fu X, Chen G, Cai ZD, et al. Overexpression of BUB1B contributes to progression of prostate cancer and predicts poor outcome in patients with prostate cancer. Onco Targets Ther 2016;9:2211-20. [PubMed]
- Cirak Y, Furuncuoglu Y, Yapicier O, et al. Predictive and prognostic values of BubR1 and synuclein-gamma expression in breast cancer. Int J Clin Exp Pathol 2015;8:5345-53. [PubMed]
- Han JY, Han YK, Park GY, et al. Bub1 is required for maintaining cancer stem cells in breast cancer cell lines. Sci Rep 2015;5:15993. [Crossref] [PubMed]
- Mur P, De Voer RM, Olivera-Salguero R, et al. Germline mutations in the spindle assembly checkpoint genes BUB1 and BUB3 are infrequent in familial colorectal cancer and polyposis. Mol Cancer 2018;17:23. [Crossref] [PubMed]
- Li L, Lei Q, Zhang S, et al. Screening and identification of key biomarkers in hepatocellular carcinoma: Evidence from bioinformatic analysis. Oncol Rep 2017;38:2607-18. [Crossref] [PubMed]
- Jeganathan K, Malureanu L, Baker DJ, et al. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J Cell Biol 2007;179:255-67. [Crossref] [PubMed]
- Jelluma N, Brenkman AB, van den Broek NJ, et al. Mps1 phosphorylates Borealin to control Aurora B activity and chromosome alignment. Cell 2008;132:233-46. [Crossref] [PubMed]
- Xie Y, Wang A, Lin J, et al. Mps1/TTK: a novel target and biomarker for cancer. J Drug Target 2017;25:112-8. [Crossref] [PubMed]
- Agarwal R, Gonzalez-Angulo AM, Myhre S, et al. Integrative analysis of cyclin protein levels identifies cyclin b1 as a classifier and predictor of outcomes in breast cancer. Clin Cancer Res 2009;15:3654-62. [Crossref] [PubMed]
- Ding K, Li W, Zou Z, et al. CCNB1 is a prognostic biomarker for ER+ breast cancer. Med Hypotheses 2014;83:359-64. [Crossref] [PubMed]
- Niméus-Malmström E, Koliadi A, Ahlin C, et al. Cyclin B1 is a prognostic proliferation marker with a high reproducibility in a population-based lymph node negative breast cancer cohort. Int J Cancer 2010;127:961-7. [Crossref] [PubMed]
- Engeland K. Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM. Cell Death Differ 2018;25:114-32. [Crossref] [PubMed]
- Uhlen M, Oksvold P, Fagerberg L, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 2010;28:1248-50. [Crossref] [PubMed]
- Sabbaghi M, Gil-Gomez G, Guardia C, et al. Defective Cyclin B1 Induction in Trastuzumab-emtansine (T-DM1) Acquired Resistance in HER2-positive Breast Cancer. Clin Cancer Res 2017;23:7006-19. [Crossref] [PubMed]
- Arsic N, Bendris N, Peter M, et al. A novel function for Cyclin A2: control of cell invasion via RhoA signaling. J Cell Biol 2012;196:147-62. [Crossref] [PubMed]
- Pagano M, Pepperkok R, Verde F, et al. Cyclin A is required at two points in the human cell cycle. EMBO J 1992;11:961-71. [Crossref] [PubMed]
- Wang G, Jiang Q, Zhang C. The role of mitotic kinases in coupling the centrosome cycle with the assembly of the mitotic spindle. J Cell Sci 2014;127:4111-22. [Crossref] [PubMed]
- Wang Q, Su L, Liu N, et al. Cyclin dependent kinase 1 inhibitors: a review of recent progress. Curr Med Chem 2011;18:2025-43. [Crossref] [PubMed]
- Westbrook L, Manuvakhova M, Kern FG, et al. Cks1 regulates cdk1 expression: a novel role during mitotic entry in breast cancer cells. Cancer Res 2007;67:11393-401. [Crossref] [PubMed]
- Pomerening JR, Ubersax JA, Ferrell JE Jr. Rapid cycling and precocious termination of G1 phase in cells expressing CDK1AF. Mol Biol Cell 2008;19:3426-41. [Crossref] [PubMed]
- Odle TG. Precision Medicine in Breast Cancer. Radiol Technol 2017;88:401M-421M. [PubMed]
- Parry D, Guzi T, Shanahan F, et al. Dinaciclib (SCH 727965), a novel and potent cyclin-dependent kinase inhibitor. Mol Cancer Ther 2010;9:2344-53. [Crossref] [PubMed]
- Cuylen S, Blaukopf C, Politi AZ, et al. Ki-67 acts as a biological surfactant to disperse mitotic chromosomes. Nature 2016;535:308-12. [Crossref] [PubMed]
- Sun X, Kaufman PD. Ki-67: more than a proliferation marker. Chromosoma 2018;127:175-86. [Crossref] [PubMed]
- Penault-Llorca F, Radosevic-Robin N. Ki67 assessment in breast cancer: an update. Pathology 2017;49:166-71. [Crossref] [PubMed]
- Lee S, Jung SR, Heo K, et al. DNA cleavage and opening reactions of human topoisomerase IIalpha are regulated via Mg2+-mediated dynamic bending of gate-DNA. Proc Natl Acad Sci U S A 2012;109:2925-30. [Crossref] [PubMed]
- Zhang R, Xu J, Zhao J, et al. Proliferation and invasion of colon cancer cells are suppressed by knockdown of TOP2A. J Cell Biochem 2018;119:7256-63. [Crossref] [PubMed]
(English Language Editor: J. Jones)


