
Expression of long-form prolactin receptor is associated with lower disease-free and overall survival in node-negative breast cancer patients
IntroductionOther Section
Breast cancer is the most frequent malignancy in women worldwide (1). The incidence of this disease is increasing in Thailand and it is also the leading cause of cancer mortality (2). Normal breast development and carcinogenesis of breast cancer are controlled by several proteins. The key modulators of mammary epithelial cell proliferation and differentiation are prolactin (PRL) and prolactin receptor (PRLR) (3).
PRLR belongs to class I cytokine receptor family. PRLR locates on chromosome 5 (p13–14) and contains at least 10 exons with overall length of over 100 kilobases (4) Alternative mRNA splicing results in five different membrane bound isoforms and proteolytic cleavage of extracellular domain (ECD) results in soluble form of PRLR (4) The membrane-bound forms comprise of long-form (LF-PRLR), intermediate-form, ΔS1-form and two short-forms (SF-PRLR). All of these isoforms, so called all-PRLR, share common ECD but different in their intracellular domain (ICD) (5). LF-PRLR, an 80–85 kDa-peptide, plays the important role in PRLR signaling via activation of several down-stream signaling pathways including JAK2, STAT5 MAPK, and PI3K, then subsequently promotes cell proliferation, differentiation, as well as survival. LF-PRLR is believed to exert the major role in PRL signaling (6). Intermediate-isoform PRLR activates JAK/STAT pathway and results in cell survival but not proliferation (7). In contrast, SF-PRLR inhibits signal transduction of MAPK (8).
PRLR has been implicated in the pathogenesis of breast and also prostate cancer (9). RT-PCR and immunohistochemistry assays showed that PRLR mRNA and protein were highly expressed in breast cancer tissue when compared to normal breast tissue (10,11). The binding of PRL to PRLR results in stimulation of breast cancer cell proliferation, migration, and survival (5). Co-expression of PRL and PRLR in breast cancer cells create autocrine/paracrine growth and survival loop (12). However, the role of PRL/PRLR signaling in breast cancer is still controversial. The absence of PRLR expression was associated with poorly-differentiated and larger tumor (13). High expression of LF-PRLR was associated with well to moderately differentiated carcinoma, negative lymph node, and prolonged metastasis-free survival (14). In contrast, specific knock down of LF-PRLR in mice model resulted in inhibition of lung metastasis. Furthermore, in vitro knock down of the LF-PRLR inhibited colony formation and induced apoptosis of the tumor cells. This evidence indicated the role of LF-PRLR in promoting breast cancer metastasis (15).
Since the role of LF-PRLR in breast cancer biology is relatively unknown, the objective of this study is to explore the associations between all-PRLR and LF-PRLR with clinicopathological parameters and survival outcome in different subtypes of breast cancers. We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/gs-20-569).
MethodsOther Section
Patients
Breast cancer patients were retrospectively recruited from the Division of Head Neck and Breast Surgery, Department of Surgery, Faculty of Medicine, Siriraj Hospital, Mahidol University, Thailand from January 2009 to December 2015. The last follow-up date was December, 31, 2019. The median follow-up time was 65.2 (6 to 158) months. The inclusion criteria are female patients with invasive ductal carcinoma, stage I–III, and age at diagnosis at 18 years old or older. All patients received standard treatment according to National Comprehensive Cancer Network guideline for treatment of breast cancer (the most recent version at the time that the patients received treatment). The exclusion criteria are presence of other primary cancer and received incomplete treatment. The data of the patients was retrieved by medical record review. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the institutional review board of Faculty of Medicine Siriraj Hospital, Mahidol University (COA. No. Si177/2016). No informed consent was obtained from the patients due to a retrospective recruitment of repository formalin-fixed paraffin-embedded tissue. The total number of the patients recruited in this study was 340.
Tissue microarray (TMA)
Formalin-fixed paraffin-embedded breast cancer tissues in excess of standard pathological examination were stored at 25 °C. TMAs were constructed manually. The TMAs were constructed by 2-mm core and composed of 3 cores per patient. The area of invasive ductal carcinoma was selected according to H&E stained slide. The cancerous area without necrosis was selected for each core, punched by 2-mm punch needle, and then placed into the recipient mold. The mold was melted at 60 °C for 6 minutes and re-embedded with new paraffin. T47D breast cancer cell line that expresses high levels of PRLR was used as the positive control for all-PRLR and LF-PRLR antibodies. The cells were collected by standard trypsinization method and centrifuged at 2,000 rpm for 5 minutes. Supernatant was removed and the cells were fixed by addition of neutral buffered formalin for 60 minutes. The cell button was then embedded in paraffin by standard protocol.
Immunohistochemical staining
The TMA sections were baked overnight at 60 °C followed by deparaffinization and rehydration. Antigen retrieval was performed by digestion with Proteinase K (Agilent Technologies, CA, USA) for 10 minutes at room temperature. Non-specific antigens were blocked by 3% hydrogen peroxide for 10 minutes. The sections were incubated with two different primary PRLR antibodies. B6.2 monoclonal antibody (Thermo Fisher Scientific, MA, USA, dilution 1:100, 32 minutes) that specific for ECD of PRLR binds to all-PRLR while H-300 polyclonal antibody (Santa Cruz Biotechnology, TX, USA, dilution 1:150, 60 minutes) that specific for ICD of PRLR binds to LF-PRLR. Then, the sections were incubated with HRP-conjugated secondary antibody (Ventana Medical Systems, AZ, USA) for 60 minutes and was visualized by ultraVIEW Universal DAB Detection KitTM (Ventana Medical Systems, AZ, USA). The sections were then counterstained with hematoxylin for 10 seconds.
Immunohistochemical scoring
The staining intensity of all-PRLR and LF-PRLR was classified as negative, weak, moderate, and strong. The expressions of all-PRLR and LF-PRLR were scored according to the percentage of positive staining cell exclusively. Negative expression was defined as absence or presence of less than 10% of cancer cell staining. Low expression was defined as 10% to <50% of cancer cell staining. High expression was defined as ≥50% of cancer cell staining. The sections were evaluated independently by two experienced pathologists who were blinded to the patients’ data. The average percentage of 3 cores and the strongest intensity of cancer cell staining were used as representative of each case.
Statistical analysis
The sample size was calculated using the formula for case-control study (16). The parameters included in the formula were: proportion of exposure in case group =0.15; proportion of exposure in control group =0.31; ratio =1; alpha =0.05, and beta =0.2, according to the results by Hachim et al. (14). The calculated sample size was 108 for each group (total =216). Chi-Square statistic was used to determine the associations between categorical variables. Odds ratio (OR) and 95% confidence interval (CI) were calculated by binary logistic regression. Disease-free survival (DFS) and overall survival (OS) were determined by Kaplan-Meier method. DFS was the time from date of surgery to time of event occurred (recurrent, metastasis, breast cancer related death). OS was the time from date of surgery to the time of death of any cause. Log-rank test was used to evaluate the significant difference of DFS and OS among different parameters. Multivariate analysis was analyzed by Cox regression. All of the statistical analysis was performed by SPSS statistics version 21 (IBM Corp., NY, USA). The statistically significant level was defined as P value <0.05.
ResultsOther Section
Characteristics of breast cancer patients
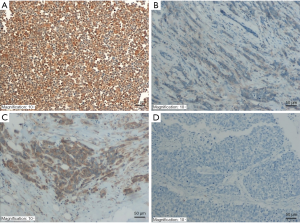
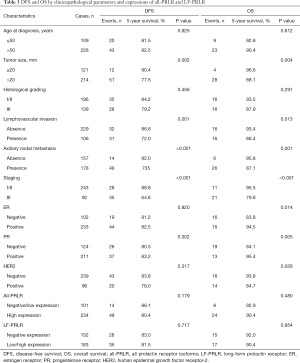
A total of 340 patients with invasive ductal carcinoma were included in this study. The mean age at diagnosis was 54.3±10.8 years. Half of the patients had stage II breast cancer. Luminal B human epidermal growth factor receptor-2 (HER2) negative is the most common subtype accounting for 47.6%. Table 1 summarized the clinicopathological parameters of the patients.
Full table
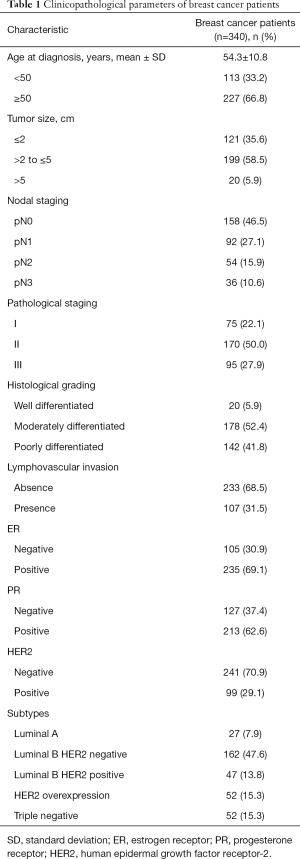
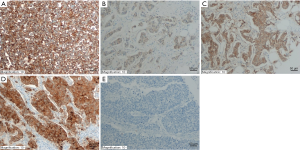
Immunohistochemical staining of all-PRLR and LF-PRLR in breast cancer tissues
Representative immunohistochemical staining of all-PRLR and LF-PRLR was showed in Figures 1,2. Both all-PRLR and LF-PRLR staining showed membranous and/or granular cytoplasmic staining in cancer cells. Positive all-PRLR staining can be classified as weak, moderate, and strong intensity, whereas there was no LF-PRLR strong intensity staining identified in the recruited patients. The majority of the patients had positive all-PRLR expression (86.2%). According to the classification by percentage of stained cells, 47 (13.8%), 56 (16.5%), and 237 (69.7%) patients had negative, low, and high all-PRLR expression, respectively. In contrast, 54.4% of the patients had positive LF-PRLR expression. The numbers of patients with negative, low and high LF-PRLR expression were 155 (45.6%), 75 (22.1%), and 110 (32.4%), respectively.
The association between all-PRLR /LF-PRLR expression and clinicopathological parameters
All-PRLR was frequently co-expressed with estrogen receptor (ER) and progesterone receptor (PR) (P<0.001). High all-PRLR expression was inversely associated with poorly differentiated carcinoma (OR: 0.402, 95% CI: 0.250–0.645). In addition, there was a trend toward presence of lymphovascular invasion and presence of axillary nodal metastasis in the patients with positive all-PRLR, however, this was not significant. LF-PRLR was also frequently co-expressed with ER and PR. The expression of LF-PRLR was associated with poorly differentiated carcinoma (OR: 1.784, 95% CI: 1.149–2.770) and HER2-overexpressed breast cancer (OR: 2.644, 95% CI: 1.602–4.366). The distribution of all-PRLR and LF-PRLR expressions among different clinicopathological parameters were summarized in Table 2.
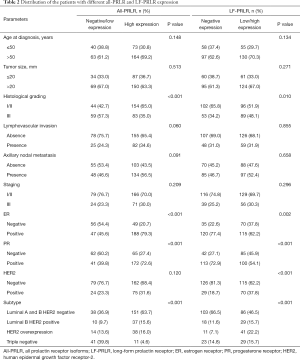
Full table
Survival analysis
A total of 335 patients with complete follow-up data were included in the survival analysis. During follow-up period, there were 17 locoregional recurrence, 44 distant metastasis, and 32 death. Survival analysis by log-rank test showed that the patients with tumor size larger than 2 cm, lymphovascular invasion, axillary nodal metastasis, and stage III had significantly lower DFS. The patients with tumor size larger than 2 cm, lymphovascular invasion, axillary nodal metastasis, stage III, negative ER, negative PR, and positive HER2 status had significantly lower OS. The expressions of all-PRLR and LF-PRLR were not associated with DFS or OS (Table 3).
Full table
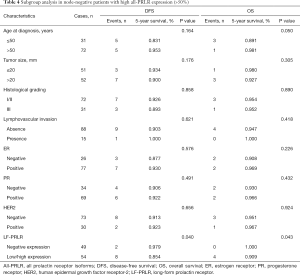
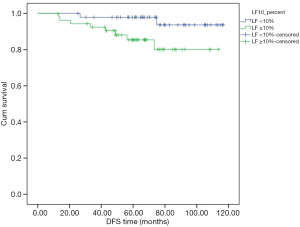
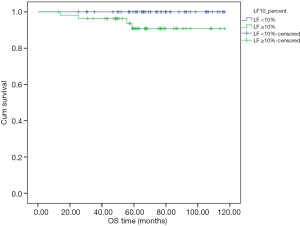
Subgroup analysis in node-negative patients with high expression of all-PRLR showed that the one with expression of LF-PRLR had significant lower DFS and OS (P=0.040 and 0.043, respectively) (Table 4). Figures 3,4 demonstrated the Kaplan-Meier curves of DFS and OS in this subgroup of patients comparing between different LF-PRLR status. Multivariate analysis by Cox regression using the factors that had P value <0.2 by log-rank test showed that LF-PRLR expression was the independent predictor for lower DFS (Table 5). However, there was no significant difference in OS between different LF-PRLR status on multivariate analysis (data not shown).
Full table

Full table
DiscussionOther Section
Several studies of association between PRLR and clinicopathological outcomes of breast cancer have been limited due to different methods of detection and lack of standard scoring system. Therefore, we set out to determine the differential expression of PRLR isoforms, LF-PRLR, in particular, and the association with breast cancer aggressiveness. We performed immunohistochemical staining using two primary antibodies to detect different isoforms of PRLR that stimulate particular downstream signaling pathways.
All-PRLR expression was found in the majority of breast cancer according to previous study (11,13). Several immunohistochemical studies of PRLR in normal, benign, and cancerous breast tissue revealed different results due to lack of standard PRLR immunohistochemical staining protocol and scoring system (11,14). A study by Gill et al. using B6.2 antibody that binds ECD of all-PRLR showed that all-PRLR was expressed in all normal breast tissue with various intensity, however, the expression was seen in the luminal borders of the epithelial cells. In invasive breast carcinoma, all-PRLR expression was seen in more than two-thirds of the patients and the staining was mostly cytoplasmic (11).
In the current study, co-expression of ER/PR and PRLR was observed in accordance with previous reports (11,17). Increased PRLR expression was also observed in luminal breast cancer cell lines treated with estrogen (17). PRL induces expression of PRLR via the recruitment of ER and STAT5 onto the promoter of PRLR (18). We also found that all-PRLR expression was associated with well/moderately differentiated tumor. The finding was concordant with a study by Faupel-Badger et al., that used antibody specific for ECD of all-PRLR. They found that negative or low all-PRLR expression was associated with larger tumor size and poorly differentiated tumor (13). In contrast, LF-PRLR expression was associated with poorly differentiated carcinoma and HER2 overexpression. This finding was discordant with the study by Hachim et al. that used H-300 antibody which is specific for LF-PRLR. They found that the expression of LF-PRLR is downregulated in invasive breast cancer and the expression was associated with favorable clinicopathological parameters (14). However, the staining was performed in a commercially purchased TMA that incorporated a small number of patients (84 invasive carcinomas).
The association between LF-PRLR and lower DFS can be demonstrated in a subgroup of node-negative breast cancer patients with high all-PRLR expression. The significance of LF-PRLR expression cannot be demonstrated regardless of axillary nodal status. This finding might be due to the impact of axillary nodal involvement on breast cancer survival (19).
LF-PRLR exerts signaling capacities via intra-cellular domain that acts as signaling unit of the receptor and contains Box 1 and 2 motifs (20). Box 1 region mediates binding to tyrosine kinase and Janus kinase (JAK2) (21). The distal residue is crucial for STAT5 binding and activation (22). LF-PRLR promotes cell proliferation, differentiation, and cell survival in breast cancer. Homodimerization of LF-PRLR activates phosphorylation of JAK2 and STAT5 and subsequently transcription of target genes while heterodimerization of LF-PRLR and SF-PRLR or homodimerization of SF-PRLR activates phosphorylation of JAK2 but did not lead to the transcription (23). Neutralization of PRLR by monoclonal antibody resulted in reduction of phosphorylation of STAT5, AKT, and ERK. In addition, this neutralizing antibody inhibited tumor growth in mice model (24). Evaluation of different PRLR isoforms might be beneficial for selection of the patients who will receive neutralizing monoclonal antibody.
In the current study, 70% of the patients with HER2 overexpression had LF-PRLR expression. To the best of our knowledge, this is the first report that found the co-expression of LF-PRLR and HER2. The possible mechanism might be explained by the transactivation between PRL/PRLR and HER2 signaling. Autocrine secretion of PRL can constitutively stimulate MAPK in breast cancer cells overexpressing HER2 via activation of JAK2 kinase and blockade of PRL/PRLR signaling reduced the levels of tyrosine phosphorylation of HER2 (25). Combination of trastuzumab and PRLR agonist showed additive inhibitory effect on MAPK phosphorylation and inhibition of cell proliferation (26). Combined blockade of PRL/PRLR and HER2 signaling might be an alternative modality for this group of patients who had anti-HER2 resistance.
When compared to other breast cancer subtypes, PRLR expression was downregulated in triple-negative breast cancer. This finding was in accordance with previous study by López-Ozuna et al. that reported downregulation of PRLR in triple-negative breast cancer (27). PRL/PRLR signaling pathway was associated with favorable survival outcome in triple-negative breast cancer (27,28). Restoration of PRL/PRLR signaling pathway in triple-negative breast cancer cell line can reduce cell viability and aggressiveness of the cancer cells (27). However, there was no association between PRLR expression in triple-negative subtype and clinicopathological outcomes in our current study due to small number of triple-negative breast cancer patients. Moreover, targeting PRL/PRLR pathway for breast cancer treatment might be benefit in a small proportion of triple-negative breast cancer patients due to low expression of PRLR in this subtype.
Limitations of this study include the recruitment of heterogeneous breast cancer population that results in unequal distribution of breast cancer in particular subtypes. Further study that determine the significance of LF-PRLR in HER2-overexpressed breast cancer might lead to the novel combined targeted therapy for HER2-overexpressed breast cancer patients.
ConclusionsOther Section
We demonstrated in a single institution breast cancer cohort that expression of all-PRLR and LF-PRLR in early breast cancer was associated with unfavorable survival. LF-PRLR expression was associated with poorly differentiated and HER2 overexpression breast cancer. LF-PRLR was the independent predictive factor for lower DFS in node-negative breast cancer patients. We proposed that LF-PRLR may be used as a potential target for breast cancer treatment.
AcknowledgmentsOther Section
The authors would like to thank Miss Wanee Pongthong (affiliation: Department of Anatomy, Faculty of Medicine Siriraj Hospital, Mahidol University), Miss Chollada Thasripoo (affiliation: Department of Immunology, Faculty of Medicine, Siriraj Hospital, Mahidol University), Miss Surat Phumphuang, and Miss Salida Ali (affiliation: Division of Head Neck and Breast Surgery, Department of Surgery, Faculty of Medicine, Siriraj Hospital, Mahidol University) for helping with data collection and manuscript preparation.
Funding: This study was supported by Siriraj Graduate Scholarship and Siriraj Grant for Research Development and Medical Education, Faculty of Medicine Siriraj Hospital, Mahidol University (Grant No. R016133002).
FootnoteOther Section
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/gs-20-569
Data Sharing Statement: Available at http://dx.doi.org/10.21037/gs-20-569
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/gs-20-569). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethic committee of Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand (COA. No. Si177/2016) and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
ReferencesOther Section
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Sa-Nguanraksa D, Sasanakietkul T. Gail model underestimates breast cancer risk in Thai population. Asian Pac J Cancer Prev 2019;20:2385-9. [Crossref] [PubMed]
- Freeman ME, Kanyicska B, Lerant A, et al. Prolactin: structure, function, and regulation of secretion. Physiol Rev 2000;80:1523-631. [Crossref] [PubMed]
- Abramicheva PA, Smirnova OV. Prolactin receptor isoforms as the basis of tissue-specific action of prolactin in the norm and pathology. Biochemistry (Mosc) 2019;84:329-45. [Crossref] [PubMed]
- Clevenger CV, Furth PA, Hankinson SE, et al. The role of prolactin in mammary carcinoma. Endocr Rev 2003;24:1-27. [Crossref] [PubMed]
- Bouilly J, Sonigo C, Auffret J, et al. Prolactin signaling mechanisms in ovary. Mol Cell Endocrinol 2012;356:80-7. [Crossref] [PubMed]
- Ben-Jonathan N, LaPensee CR, LaPensee EW. What can we learn from rodents about prolactin in humans? Endocr Rev 2008;29:1-41. [Crossref] [PubMed]
- Devi YS, Seibold AM, Shehu A, et al. Inhibition of MAPK by prolactin signaling through the short form of its receptor in the ovary and decidua: involvement of a novel phosphatase. J Biol Chem 2011;286:7609-18. [Crossref] [PubMed]
- Goffin V. Prolactin receptor targeting in breast and prostate cancers: New insights into an old challenge. Pharmacol Ther 2017;179:111-26. [Crossref] [PubMed]
- Touraine P, Martini JF, Zafrani B, et al. Increased expression of prolactin receptor gene assessed by quantitative polymerase chain reaction in human breast tumors versus normal breast tissues. J Clin Endocrinol Metab 1998;83:667-74. [Crossref] [PubMed]
- Gill S, Peston D, Vonderhaar BK, et al. Expression of prolactin receptors in normal, benign, and malignant breast tissue: an immunohistological study. J Clin Pathol 2001;54:956-60. [Crossref] [PubMed]
- Reynolds C, Montone KT, Powell CM, et al. Expression of prolactin and its receptor in human breast carcinoma. Endocrinology 1997;138:5555-60. [Crossref] [PubMed]
- Faupel-Badger JM, Duggan MA, Sherman ME, et al. Prolactin receptor expression and breast cancer: relationships with tumor characteristics among pre- and post-menopausal women in a population-based case-control study from Poland. Horm Cancer 2014;5:42-50. [Crossref] [PubMed]
- Hachim IY, Hachim MY, Lopez VM, et al. Prolactin receptor expression is an independent favorable prognostic marker in human breast cancer. Appl Immunohistochem Mol Morphol 2016;24:238-45. [Crossref] [PubMed]
- Yonezawa T, Chen KH, Ghosh MK, et al. Anti-metastatic outcome of isoform-specific prolactin receptor targeting in breast cancer. Cancer Lett 2015;366:84-92. [Crossref] [PubMed]
- Rosner B. Fundamentals of biostatistics. 5th ed. Pacific Grove: Duxbury; 2000:792.
- Ormandy CJ, Hall RE, Manning DL, et al. Coexpression and cross-regulation of the prolactin receptor and sex steroid hormone receptors in breast cancer. J Clin Endocrinol Metab 1997;82:3692-9. [Crossref] [PubMed]
- Kavarthapu R, Tsai Morris CH, Dufau ML. Prolactin induces up-regulation of its cognate receptor in breast cancer cells via transcriptional activation of its generic promoter by cross-talk between ERalpha and STAT5. Oncotarget 2014;5:9079-91. [Crossref] [PubMed]
- Hortobagyi GN, Edge SB, Giuliano A. New and important changes in the TNM staging system for breast cancer. Am Soc Clin Oncol Educ Book 2018;38:457-67. [Crossref] [PubMed]
- Chilton BS, Hewetson A. Prolactin and growth hormone signaling. Curr Top Dev Biol 2005;68:1-23. [Crossref] [PubMed]
- Pezet A, Buteau H, Kelly PA, et al. The last proline of Box 1 is essential for association with JAK2 and functional activation of the prolactin receptor. Mol Cell Endocrinol 1997;129:199-208. [Crossref] [PubMed]
- Pezet A, Ferrag F, Kelly PA, et al. Tyrosine docking sites of the rat prolactin receptor required for association and activation of stat5. J Biol Chem 1997;272:25043-50. [Crossref] [PubMed]
- Kang JH, Hassan SA, Zhao P, et al. Impact of subdomain D1 of the short form S1b of the human prolactin receptor on its inhibitory action on the function of the long form of the receptor induced by prolactin. Biochim Biophys Acta 2014;1840:2272-80. [Crossref] [PubMed]
- Damiano JS, Rendahl KG, Karim C, et al. Neutralization of prolactin receptor function by monoclonal antibody LFA102, a novel potential therapeutic for the treatment of breast cancer. Mol Cancer Ther 2013;12:295-305. [Crossref] [PubMed]
- Yamauchi T, Yamauchi N, Ueki K, et al. Constitutive tyrosine phosphorylation of ErbB-2 via Jak2 by autocrine secretion of prolactin in human breast cancer. J Biol Chem 2000;275:33937-44. [Crossref] [PubMed]
- Scotti ML, Langenheim JF, Tomblyn S, et al. Additive effects of a prolactin receptor antagonist, G129R, and herceptin on inhibition of HER2-overexpressing breast cancer cells. Breast Cancer Res Treat 2008;111:241-50. [Crossref] [PubMed]
- López-Ozuna VM, Hachim IY, Hachim MY, et al. Prolactin pro-differentiation pathway in triple negative breast cancer: impact on prognosis and potential therapy. Sci Rep 2016;6:30934. [Crossref] [PubMed]
- Motamedi B, Rafiee-Pour HA, Khosravi MR, et al. Prolactin receptor expression as a novel prognostic biomarker for triple negative breast cancer patients. Ann Diagn Pathol 2020;46:151507. [Crossref] [PubMed]



