
Predicting the likelihood of early recurrence based on mRNA sequencing of pituitary adenomas
Introduction
Pituitary adenoma (PA), which arises from adenohypophysis cells, is the second most common intracranial tumor with a prevalence of 1/500 (1). Based on tumor diameter, the PA is classified as a microadenoma, macroadenoma, or giant adenoma. The recurrence rate of incidental PA is 9.6% (2). Lv et al. reported that tumor size and radiological residuals are risk factors for PA recurrence (3). However, many patients experience recurrent tumors after surgery in spite of small tumor size and no radiological residuals, indicating that tumor properties also play a significant role in tumor recurrence, leading to the investigation of genetic factors that drive the tumor property of recurrence.
Although patients with a giant tumor are more likely to experience tumor recurrence (4), not all giant pituitary tumors are associated with tumor recurrence, as many microadenomas and macroadenomas with or without residuals also share the risk of tumor recurrence (5). However, clinical function status, radiological features, and histology cannot reliably and accurately predict tumor recurrence (6,7). Therefore, the ability to predict tumor recurrence in patients with PA is valuable.
With the progress of next-generation sequencing technology over the last decade, comprehensive interpretation of tumor biology has been widely deployed in various neoplastic diseases (8). Transcriptome sequencing, especially mRNA-Seq, has a relatively low cost and provides precise insight into the complex transcriptome of tumors. By combining the most advanced technology and a detailed follow-up of patients, we aimed to predict early recurrence in PA patients, which may help to improve the prognostic assessment of PAs, resulting in treatment options for high-risk patients to lower the potential of recurrence.
Methods
Patients and samples
We obtained the records and samples from 107 patients who underwent surgery for PA at the Beijing Tiantan Hospital between 2014 and 2016. Studies on human material were approved by the Institution Research Ethics Committee (approval number KYSB2017-142-01). Appropriate consent for the use of the material was acquired from patients enrolled in the study.
mRNA-Seq data
A total of 107 freshly frozen PA samples were collected for the initial exploratory analysis using RNA-Seq. The demographics of this patient population are shown in Table 1. Central pathology reviews of these specimens were based on 2017 WHO classification (9). All samples collected contained at least 80% viable PA tissue, and they were frozen within 5 min after resection until utilized for RNA-Seq analysis. In total, 16,714 genes were selected to construct the model, and only protein-coding genes that were expressed in over 50% of all samples were included.
Full table
Definition of early recurrence
Early recurrence was defined as a return of abnormal plasma hormone levels and/or radiological evidence of a significant increase of tumor volume within 24 months post-operation. The tumor volume was decided by measuring the largest tumor diameter by 3 dimensions (x, y, and z), and the volume was estimated using the following formula: 1/3πxyz. The criterion used to characterize a significant increase in tumor volume was a 20% increase in size compared to the previous tumor. Any visible abnormal tumor-like signal in MRI imaging 3 months post-operation was defined as radiological residual, and any abnormal serum hormone level 3 months post-operation was defined as hormone recurrence.
Patients who had no evidence of disease (no clinical symptoms, normal plasma hormone levels, and no visible radiological rumor remnant) 24 months post-operation were considered as recurrence-free. The follow-up started one-year after surgery. Tumors recurring later than 24 months were assigned to the early recurrence-free group.
Model
In total, 107 samples with both mRNA-Seq data and postoperative follow-up data were obtained in this study. All samples were randomly divided into the following two groups: a training set (n=57) and a validation set (n=50). Cox and logistic regression analyses were applied to predict the presence of a certain outcome based on available variables, which has been successfully used for prediction and diagnosis in medicine (10,11). In the training group, the log2-normalized expression value of the qualified genes (n=16,714) and the recurrence status were applied to the Cox regression. The genes were then ranked according to the P value of Cox regression. The risk score was introduced to quantify the risk of tumor early recurrence, and it comprised the summation of the product of the top n gene expression values and corresponding β-values from Cox regression as follows:
A receiver operating characteristic (ROC) curve was obtained along with the area under the curve (AUC) of the top n gene to screen for genes with the best prediction value. Considering the number of genes and AUC, the top 7 genes in the module that approached the plateau were used to construct the prediction model. The risk score of every individual was calculated according to the formula above, and the cutoff value was defined by calculating the Youden index (cutoff value =10.78).
Results
Sample characteristics
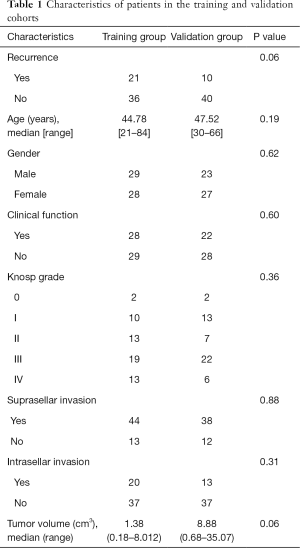
In total, 107 PA patients were randomly assigned to training (n=57) and validation (n=50) groups. The training group had 28 females and 29 males with a median age of 44.78 years old (range, 21–84 years). The validation group had 27 females and 23 males with a median age of 47.52 years old (range, 30–66 years). There were 21 (37%) and 10 (20%) patients suffering from early recurrence in the training and validation groups, respectively. The patients were classified according to the Knosp grades were as follows: 10 (17.5%) and 13 (26%) patients were grades I in the training and validation groups, respectively; 13 (22.8%) and 7 (14%) patients were grade II in the training and validation groups, respectively; 19 (33.3%) and 22 (44%) were grade III in the training and validation groups, respectively; and 13 (22.8%) and 6 (12%) were grade IV in the training and validation groups, respectively. There were 28 (49.1%) and 22 (44%) patients with functional PAs in the training and validation groups, respectively. Suprasellar invasion presented in 44 (77.2%) and 38 (76%) patients in the training and validation groups, respectively. Infrasellar invasion presented in 20 (35.1%) and 13 (26%) patients in the training and validation groups, respectively. The median tumor volume was 1.38 (range, 0.18–8.01) cm3 in the training group and 8.88 (range, 0.68–35.07) cm3 in the validation group.
Significant differences were not found in the incidence of recurrence, age, gender, clinical function, Knosp grade, suprasellar invasion, intrasellar invasion, and tumor volume between the training and validations groups (Table 1).
Distinct expression profile of early recurrent samples
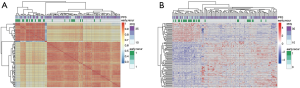
To explore the gene property of tumor early recurrence, mRNA-seq data of these 107 samples were applied. Differential expression for early recurrence was analyzed with a two-tailed t-test (P<0.05) and ranked by the relative expression fold change, and the top 100-fold change genes were selected to represent the distinct gene expression pattern. The top 100 differentially expressed genes were clustered, which indicated two distinct groups (Figure 1A), showing high consistency with early recurrence (Fisher’s exact test, P=0.0002). To further explore the similarity of these samples, the clustering of the correlation coefficients of every two samples also resulted in two distinct groups (Figure 1B). The two groups showed similar consistency with early recurrence (P<0.001). These results showed that patients with early recurrence harbored a distinct expression profile. The expression differences laid the foundation to construct a prediction model based on mRNA expression.
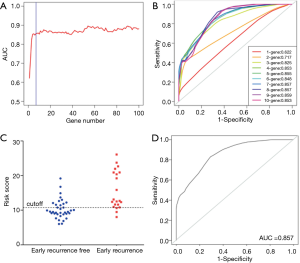
Model construction in the training set
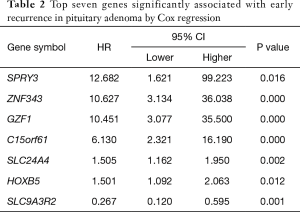
The risk score method generated the top 7 ranked genes after the Cox regression reached the plateau of the AUC (Figure 2A,B), showing the best performance of early recurrence prediction. The top 7 genes are listed in Table 2. By applying the model in the training group, 57 samples were stratified into positive and negative groups (Figure 2C), which was in high accordance with the follow-up data. This model generated an AUC of 0.857 (95% CI: 0.714–0.937), a sensitivity of 85.7%, and a specificity of 69.4% in the training set (Figure 2D).
Full table
Model evaluation in the validation set
To evaluate the predictive performance of this gene set used in the training group, the risk score generated by these 7 genes was obtained from the 50 samples in the validation set. All patients (sensitivity =100%) with early recurrence were predicted correctly by this model, while 15/40 (specificity =32.5%) patients were predicted to be negative (Figure 3A). This model had an AUC of 0.936 (95% CI: 0.874–1; Figure 3B).
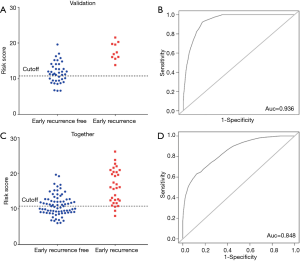
When all 107 patients were included in the Cox regression model, the following results were obtained: sensitivity of 28/31 (90.32%), the specificity of 37/76 (48.68%) (Figure 3C), and AUC of 0.848 (95% CI: 0.733–0.912; Figure 3D). The high accuracy of this model highlighted its potential diagnostic value for the early recurrence of PA.
Prognostic value of the model
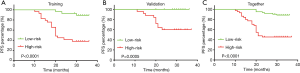
The risk scores of the patients were ranked from low to high, with the first half being assigned to the low-risk group and the second half being assigned to the high-risk group to elucidate the prognostic value of this seven-gene model. The mean follow-up after surgery in these patients was 28.0±7.0 months, and 31 patients (30.0%) had an early recurrence of PA. The Kaplan-Meier progression-free survival curves showed a significant difference between the training and validation groups (P<0.0001 and P=0.0005, Figure 4A,B). When applied to all patients, the Kaplan-Meier progression-free survival curve showed a significant difference between the high- and low-risk score patients (P<0.0001, Figure 4C). Overall, 92.9% of all patients (relatively low-risk score patients) had no early recurrence with rates of 88% and 100% in the training and validation groups, respectively.
Prognostic value of single genes in a gene set
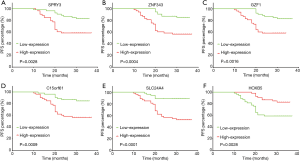
To further evaluate the prognostic feature of this gene set, we explored the relationship between recurrence and individual gene expression in this model. Two groups were separated according to the RNA expression level of each gene. The single gene expression of the 107 patients was ranked from low to high. The first half in the ranking was defined as low expression, and the second half in the ranking was defined as high expression. Kaplan-Meier progression-free survival curves were generated for each group. We found that 6 out of the 7 genes showed a significant difference between the high and low expression groups (Figure 5A,B,C,D,E,F) with P values varying from <0.0001 to 0.0028. SLC9A3R2 was the only exception with a P value of 0.0699, and it showed a trend to differentiate the prognosis (data not shown). These data indicate that the promising prognostic property may be explained by the pathways related to these genes.
Discussion
Recurrence of PA is generally accompanied by seriously devastated pituitary function and visual deficits. In addition, tumor resection for recurred tumors is more difficult than resection of primary tumors. Due to limited therapeutic options, the outcome of recurrent pituitary adenoma is still unsatisfactory. Thus, the prediction of early recurrence would provide critical information to introduce appropriate intervention to postoperative patients. Although progress in molecular biology and radiology has been made over the last few decades, the cause and hazardous factors of recurrence are still unclear, implying that a more reliable predictive method for pituitary tumor recurrence is needed. It is practical to distinguish more proliferative and aggressive adenomas by pathological markers (Ki-67, mitotic activity, and p53 staining), which has been proven to be useful in other malignancies (12). However, the lack of a standard staining technique and outcome assessment limits its prognostic value. As a result, the classification type “atypical adenoma” has been removed from the 4th edition of the WHO classification of pituitary gland tumors after years of controversy (13).
In the cases where a tumor recurred within a short period after the operation, tumor proliferation, a driving force of tumor recurrence, may be the major factor. Thus, we defined 24 months as the cutoff time. In this study, we applied an analysis based on a large number of patients over a long follow-up period. By utilizing an advanced statistical method, a prediction model based on mRNA-Seq was built to obtain the probability of early recurrence, which achieved high sensitivity and specificity. This model also indicated distinguishable characteristics in both the training and validation groups. Therefore, this model should be useful to clinicians, pathologists, and researchers.
For recurrence prediction, postoperative residuals were considered as a high-risk factor. Early recurrence occurred in 10.7% of cases with no visible postoperative residual adenoma. Of these cases, 38.8% had intrasellar residuals, and 66.7% had extrasellar residuals. The 10-year recurrence rate was 25% in patients without residual recurrences (14). In this study, the tumor early recurrence rate was 30.0%, and the mean follow-up period after surgery in these patients was 28.0±7.0 months. This result showed that the postoperative residuals and locations are individual risk factors of tumor recurrence. However, further discussion should be made to interpret the mechanism involved in the tumor property and tumor locations. Transcriptome sequencing should provide a novel, relatively low-cost insight into the complex mechanism of tumor recurrence regulated by gene expression.
In the present study, patients who suffered from an early recurrence commonly had higher risk scores. The sensitivity was 85.7% in the training group, and all patients with early recurrence were predicted correctly by this model in the validation group, leading to 100% sensitivity. Our data suggest that a screening tool combined with a proper early stage intervention, such as the transsphenoidal approach, transcranial approach surgery, and gamma knife, should be administered to patients with a high-risk score detected after primary tumor resection to decrease the overall recurrence rate. In addition, follow-up for at least two years may be required.
Even though the individual genes in our prognostic model emphasize a trend to differentiate the PA patients from higher and lower risks of early recurrence, two genes, HOXB5 and GZF1, are well-known to play a vital role in multiple molecular pathways involved in tumorigenesis and cancer-like behaviors. These two genes, along with all the other genes in this model, may have potential prognostic value and be qualified to be therapeutic targets for recurrent PAs.
HOXB5 is a member of the Antp homeobox family and encodes a nuclear protein with a homeobox DNA-bind domain. It is well-known that homeobox genes (HOX genes) are involved in tumorigenesis. A previous study has shown that HOXB5 is associated with cell migration and invasion in lung cancer, retinoblastoma, and gastric cancer (15-17), suggesting that it may be an important regulator of the Wnt/β-catenin signaling pathway or ERK1/2 (15,17). To the best of our knowledge, no study has reported the relationship between HOXB5 and pituitary tumor cell migration and invasion. Therefore, larger, prospective validations of the HOXB5 signature for its prediction values in patients with pituitary tumors are warranted, and functional and mechanistic studies on HOXB5 should be performed to further support its clinical application in pituitary tumors. Furthermore, HOXB5 may be a target oncogene, which could be inhibited by miR-1275, and result in the suppression of G1/S transition (18), thus potentially leading to a better outcome for PA patients.
GZF1 is a zinc finger protein induced by the glial cell-line-derived neurotrophic factor, which seems to play a crucial role in cell proliferation (19). We suspect that this gene may play a critical role in pituitary tumor recurrence, and further identification should be conducted. Although these seven genes have not been widely reported to be associated with pituitary cell proliferation, invasion, and recurrence, some of these genes showed a tendency to be related to cell proliferation and metastasis, which may potentially cause tumor recurrence.
Although this model was constructed based on high-throughput mRNA sequencing from 107 samples, it still needs further validation in the other datasets. Limited by the shortage of sequencing data with long-term follow-ups, the further comprehensive evaluation of the model needs to be explored in the future.
Conclusions
In summary, we established a predictive model that showed high sensitivity for PA recurrence. This is the first PA recurrence prediction model derived from mRNA-Seq data. The application of this model may help to identify the early recurrence of pituitary tumors and to administrate more appropriate therapeutic and follow-up strategies for patients with PAs.
Acknowledgments
Funding: This work was supported by grants from the Natural Science Foundation of Beijing (No. 7142054).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Studies on human material were approved by the Institution Research Ethics Committee (approval number KYSB2017-142-01). Appropriate consent for the use of the material was acquired from patients enrolled in the study.
References
- Fernandez A, Karavitaki N, Wass JA. Prevalence of pituitary adenomas: a community-based, cross-sectional study in Banbury (Oxfordshire, UK). Clin Endocrinol (Oxf) 2010;72:377-82. [Crossref] [PubMed]
- Seltzer J, Wedemeyer MA, Bonney PA, et al. Outcomes following transsphenoidal surgical management of incidental pituitary adenomas: a series of 52 patients over a 17-year period. J Neurosurg 2018. [Epub ahead of print]. [PubMed]
- Lv L, Hu Y, Yin S, et al. Clinically aggressive phenotype: A clinicopathological case series of atypical pituitary adenomas. Clin Neurol Neurosurg 2018;167:93-8. r. [Crossref] [PubMed]
- Ling X, Zhu T, Luo Z, et al. A review of pituitary abscess: our experience with surgical resection and nursing care. Transl Cancer Res 2017;6:852-9. [Crossref]
- Losa M, Mortini P, Barzaghi R, et al. Early results of surgery in patients with nonfunctioning pituitary adenoma and analysis of the risk of tumor recurrence. J Neurosurg 2008;108:525-32. [Crossref] [PubMed]
- Karki M, Sun J, Yadav CP, et al. Large and giant pituitary adenoma resection by microscopic trans-sphenoidal surgery: Surgical outcomes and complications in 123 consecutive patients. J Clin Neurosci 2017;44:310-4. [Crossref] [PubMed]
- Bacuzzi A, Dionigi G, Guzzetti L, et al. Predictive features associated with thyrotoxic storm and management. Gland Surg 2017;6:546-551. [Crossref] [PubMed]
- Doroshow JH, Kummar S. Translational research in oncology--10 years of progress and future prospects. Nat Rev Clin Oncol 2014;11:649-62. [Crossref] [PubMed]
- Inoshita N, Nishioka H. The 2017 WHO classification of pituitary adenoma: overview and comments. Brain Tumor Pathol 2018;35:51-6. [Crossref] [PubMed]
- Gareen IF, Gatsonis C. Primer on multiple regression models for diagnostic imaging research. Radiology 2003;229:305-10. [Crossref] [PubMed]
- Bagley SC, White H, Golomb BA. Logistic regression in the medical literature: standards for use and reporting, with particular attention to one medical domain. J Clin Epidemiol 2001;54:979-85. [Crossref] [PubMed]
- Criscitiello C, Disalvatore D, De Laurentiis M, et al. High Ki-67 score is indicative of a greater benefit from adjuvant chemotherapy when added to endocrine therapy in luminal B HER2 negative and node-positive breast cancer. Breast 2014;23:69-75. [Crossref] [PubMed]
- Lopes MBS. The 2017 World Health Organization classification of tumors of the pituitary gland: a summary. Acta Neuropathol 2017;134:521-35. [Crossref] [PubMed]
- Levy MJ, Robertson IJ, Khalk N, et al. Long-term follow-up of a large prospective cohort of patients with nonfunctioning pituitary adenomas: The outcome of a conservative management policy. Clin Endocrinol (Oxf) 2018;89:354-9. [Crossref] [PubMed]
- Hong CS, Jeong O, Piao Z, et al. HOXB5 induces invasion and migration through direct transcriptional up-regulation of β-catenin in human gastric carcinoma. Biochem J 2015;472:393-403. [Crossref] [PubMed]
- Xu H, Zhao H, Yu J. HOXB5 promotes retinoblastoma cell migration and invasion via ERK1/2 pathway-mediated MMPs production. Am J Transl Res 2018;10:1703-12. [PubMed]
- Zhang B, Li N, Zhang H. Knockdown of Homeobox B5 (HOXB5) Inhibits Cell Proliferation, Migration, and Invasion in Non-Small Cell Lung Cancer Cells Through Inactivation of the Wnt/β-Catenin Pathway. Oncol Res 2018;26:37-44. [Crossref] [PubMed]
- Sun KY, Peng T, Chen Z, et al. MicroRNA-1275 suppresses cell growth, and retards G1/S transition in human nasopharyngeal carcinoma by down-regulation of HOXB5. J Cell Commun Signal 2016;10:305-14. [Crossref] [PubMed]
- Dambara A, Morinaga T, Fukuda N, et al. Nucleolin modulates the subcellular localization of GDNF-inducible zinc finger protein 1 and its roles in transcription and cell proliferation. Exp Cell Res 2007;313:3755-66. [Crossref] [PubMed]


