Differential protein expressions in breast cancer between drug sensitive tissues and drug resistant tissues
Breast cancer is one of the most common malignant tumors among women, accounting for about 7-10% of all systemic malignancies. Also, its incidence is rising, and therefore has posed serious threat to women’s health. Chemotherapy plays a key role in the treatment of breast cancer; however, the widespread resistance to chemotherapy often results in the treatment failure (1,2).
Proteomics is a discipline that studies the proteome using various technical approaches. By analyzing the changes of intracellular protein components in a holistic and dynamic manner, proteomics tries to explore the mutual effects and connections among these components and finally grasps their functions and activities. By using the proteomic techniques, we can investigate the multidrug resistance of tumor cells at the protein level, and thus screen resistance-related proteins. The previous studies on multidrug resistant proteins as well as proteomics research on highly drug-resistant breast cancer cell strains could not satisfactorily explain and reserve the drug-resistance of breast cancer. Both chemotherapy-resistance-related proteins and chemotherapy-sensitivity-related proteins can be expressed in breast cancer cells. The differential expressions result in the diversity of treatment sensitivity. A combined research on both drug-resistance-related proteins and drug-sensitivity-related proteins will be helpful to elucidate the mechanism of chemotherapy resistance. Furthermore, the previous studies on drug resistance of breast cancer were conducted in breast cancer cell strains, which are somehow different from the breast cancer tissues (3,4). Therefore, in our current study, we screened chemotherapy-sensitive and chemotherapy-resistant breast cancer patients, determined the difference of protein expressions between the drug-resistant and drug-sensitive breast cancer cells using proteomic technology, and thus screen the drug resistance-related proteins.
Materials and methods
Sample collection
A total of 128 patients [aged 29-68 years; or, (45±17) years] with invasive ductal carcinoma of the breast who received neoadjuvant chemotherapy in the Second Xiangya Hospital of Central South University from May 2008 to May 2010 were enrolled in this study. Informed consents were obtained from these patients, and specimens were harvested using Mammotome biopsy and then stored frozen. TEC-based neoadjuvant chemotherapy was applied for each patient: T, taxotere (TXT), 75 mg/m2; E, epirubicin (E-ADM), 80 mg/m2; and C, cyclophosphamide (CTX), 500 mg/m2. Drugs were intravenously administered on the first day of each cycle, with 21 days as one cycle. The efficacy was evaluated after 2-4 cycles of treatment. The change of tumor size was calculated using color ultrasound, and the chemotherapy efficacy was evaluated using Response Evaluation Criteria in Solid Tumors (RECIST) (5). Finally, complete remission (CR) was achieved in 24 patients, partial remission (PR) in 53 patients, stable disease (SD) in 32 patients, progression disease (PD) in 19 patients. Ten patients were randomly selected from the CR group and PD group, respectively, as the sensitive group and resistant group.
Main instruments, reagents, and bioinformatic software
Two-dimensional protein quantification kit, acrylamide, Methylene diacrylamide, acetic acid, sodium dodecyl sulfonate (SDS), urea, cholanidopropyl dimethylammonio propanesulfonate (CHAPS), thiourea, ammonium persulfate, IPG buffer liquid (pH 3-10), 24-cm immobilized pH gradient-driven paper were puchased from Amershan Pharmacia Corporation. Dithiothreitol, iodoacetamide, trypsin, trifluoroacetic acid (TFA), ammonium bicarbonate, sodium thiosulfate, cyanide, and α-Cyano-4-hydroxycinnamic acid were purchased from Sigma, USA. The centrifuge tubes were manufactured by Millipore, USA. Phosphoric acid, glycerol, ethanol, and EDTA-Na2 were domestic products.
IPGphor isoelectric focusing system, Ettan DALT vertical electrophoresis tank, and Imagescanner scanner were produced by Amersham Biosciences, USA. Matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF-MS) equipment (Voyager-DE STR 4307), LabScan scanning software, and Data Explorer (mass spectrometry software) were purchased from Applied Biosystem, USA. PDQuest™ 7.0 gel image analysis software was purchased from Bio-Rad, USA. Mascot Distiller for mass spectrum peak identification and Mascot for peptide mass fingerprinting database query were products of Matrixscience, UK. SignalP (3.0) softwared was provided by Swiss-Prot database.
Methods
Detection using proteomic methods
Samples were ground into a fine powder under liquid nitrogen, and then put into EP tubes. The supernatants were collected after cleavage for detecting the protein concentrations. Dual gel electrophoresis was performed according to the manufacturer’s instructions (6). In each test, the fixed loading amount was 1,000 μg. In the sensitive group, the protein was mixed into 1,000 µg proportionally and then loaded onto an IPG slot containing IPG strips. In the resistant group, proteins were mixed using the same method. Isoelectric focusing was conducted according to the following procedure: 30 V × 12 h, 500 V × 1 h, 1,000 V × 1 h, and 8,000 V × 8.5 h. After equilibration, the product underwent SDS-PAGE gel electrophoresis and Coomassie brilliant blue staining. Destain until background is clear. 2-DE maps were obtained by scanning the gels. PDQuest 2-D analysis software was used for spot detection and spot matching, so as to identify the potential differences. Excise the protein gel spots of interest and destain them for reduction and alkylation. After in situ enzymatic hydrolysis, add extraction liquid, and then refrigerate and compress the samples. Obtain 0.5 μL samples, mix them with equal volume of saturated cyano-4-hydroxycinnamic acid (CHCA), and then spot them onto sample plate. The parameters were set up as following: positive ion-reflector mode, accelerating voltage 20 kV, mirror voltage ratio 1.12, N2 laser wavelength 337 nm, pulse width 3 ns, the number of laser shots 50, delay 100 nsec, and vacuum degree 4×10-7 Torr. A trypsin-fragment peak (842.510, 2211.105) was served as internal standard for mass calibration. A list of the corrected mass peaks was the peptide mass fingerprinting (PMF). Mascot Wizard software was used for database searching. Proteins were identified using the SwissProt database.
Identification of differential expressions of proteins with Western blotting
The differential expressions of the relevant proteins in the sensitive group (n=20) and resistant group (n=20) were determined using Western blotting (7).
Statistical analysis
Statistical analyses were done using SPSS 15.0 software. The means were compared using chi square test and t test. All measurement data are presented as mean ± standard deviation (χ ± SD). Rate comparison was conducted using chi square test. P
Results
Two-dimensional electrophoresis profiles in the sensitive group and resistant group
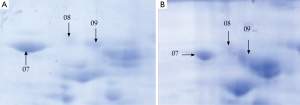
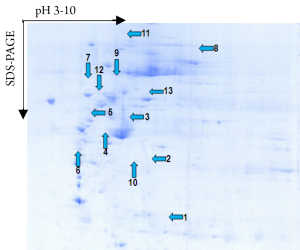
A total of 53 protein spots with expression differences larger than 2 times were obtained (26 up-regulated and 27 down-regulated in the resistant group). Some differentially expressed protein spots are shown in Figure 1. Some selected protein spots are enlarged in Figure 2.
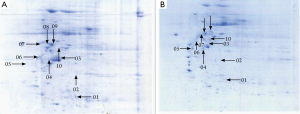
Identification of differentially expressed proteins
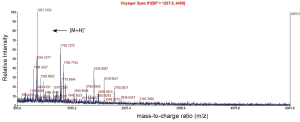
A Mascot score of >64 (PFigure 3). A selected PMF of protein spot 6 is display in Figure 4. Thirteen differentially expressed proteins were found in SWISS-PROT protein database, among which 3 up-regulated and 10 down-regulated in the resistant group (Table 1).
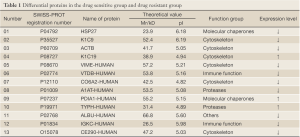
Full Table
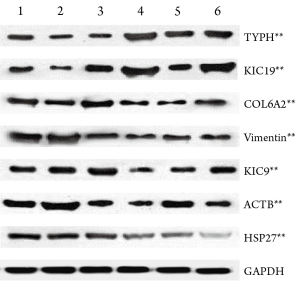
Western blot verification of mass spectrometry results
Western blot verification was performed for 7 potentially meaningful proteins, which showed that the expressions of keratin type I cytoskeletal 19 (KIC19) and thymidine phosphorylase (TYPH) were remarkably up-regulated in the resistent group, whereas those of heat shock protein 27 (HSP27), keratin cytoskeleton-I 9 (KIC9), collagen alpha-2 (VI) (CO6A2), vimentin (VIME), and beta-actin cytoplasmic 1 (ACTB) were significantly down-regulated (PFigure 5).
Discussion
In our current study, by comparing the differentially expressed proteins between chemotherapy-resistant and sensitive breast cancer cells using the two-dimensional electrophoresis technology for proteomics, we successfully obtained the two-dimensional gel electrophoresis maps with high resolution and good repeatability. Totally 53 differential protein spots were found, and 13 differentially expressed proteins were identified, among which 3 proteins up-regulated and 10 down-regulated in chemotherapy-resistant cells. Based on the results of literature review, we conducted Western plot identification for the expressions of 7 proteins that may be related with the tumor occurrence and progression as well as chemotherapy sensitivity and resistance. The results were consistent with the findings of proteomic studies: the expressions of KIC19 and TYPH were up-regulated in the resistant group, indicating that these two proteins may be related with chemotherapy resistance; in contrast, the expressions of HSP27, KIC9, CO6A2, VIME, and ACTB were down-regulated in the resistant group.
KIC19 is highly tissue-specific. It is not expressed in the normal bone marrow, peripheral blood, and lymph nodes, but is widely expressed in breast cancer cells. Therefore, KIC19 has been a key marker of the micrometastasis of breast cancer (8-10). KIC19 may be related with the intracellular and intercellular signaling, facilitating the invasion and metastasis of tumors (11). In our current study, the expression of KIC19 was up-regulated in the resistant group, suggesting that it may be associated with drug resistance.
TYPH plays a key role in the synthesis and decomposition of pyrimidine nucleosides. It can induce angiogenesis and suppress apoptosis, and therefore is closely associated with the growth and metastasis of tumors. Its main functions include: (I) reversibly catalyzes the dephosphorylation of thymidine to produce 2-Deoxyribose-D-Ribose-1-Phosphoric acid and thymine, maintaining the thymidine at a relatively stable level; (II) transfers the deoxyribose on deoxy-nucleosides to other basic groups, facilitating the formation of new deoxy-nucleosides; (III) promotes angiogenesis in vivo and stimulates the chemotaxis and growth of endotheliocytes in vitro (12); (IV) catalyzes the reaction between 1-phosphate-2-deoxyribose and fluorouracil (FU, an anti-pyrimidine drug) to produce 5-fluoro-2-deoxyuridine (5-FdUrd), and finally produces end products with anti-cancer activities. 5-deoxy-5-floxuridine and capecitabine are the precursors of FU. As a key rate-limiting enzyme that regulates their transformation into FU, TYPH can inhibit DNA synthesis and thus exert its anti-tumor effect. The mechanism governing the up-regulation of TYPH remains unclear; indeed, the up-rugulation may be induced by some cytokines. Research (13) has indicated that chemotherapy drugs (e.g., paclitaxel, cyclophosphamide, docetaxel, and mitomycin C) and radiotherapy may induce the upregulation of TYPH expression by up-regulating the gamma-interferon (INF-G), tumor necrosis factor-alpha (TNF-α), interleukin-1α (IL-1α), or IL-10. In our current study, TYPH paclitaxel chemotherapy for breast cancer significantly increased expression in resistance groups, prompting TYPH expression is likely to play a role in resistance to chemotherapy.
The expressions of other proteins including HSP27, KIC9, CO6A2, VIME, and ACTB were down-regulated in the resistant group and up-regulated in the sensitive group, suggesting that these proteins may be associated with chemotherapy sensitivity. Among them, HSP27 is a low-molecular-weight member in the heat shock protein family. Many studies (14,15) have shown that HSP27 is associated with the growth and apoptosis of cells, drug resistance, and occurrence, progression, and metastasis of tumors. In recent years, the correlation of HSP27 with chemotherapy resistance has been widely explored and remains controversial. A study (14) has shown that HSP27 could induce the hamster ovary cells become resistant to adriamycin and other related drugs. In some malignant tumors such as esophageal squamous cell carcinoma and leukemia, the HSP27 over-expression induced resistance to chemotherapy. It can alleviate the cytotoxic effects of actinomycete D, colchicine, daunorubicin, vincristine, and sodium arsenide, and thus Increase cell survival. Tumor cells with high HSP27 expression had increased resistance to Adriamycin (15). According to Seymour et al. (16), HSP27-positive metastatic breast cancer is more sensitive to chemotherapy, has better prognosis, and is associated with histological grade.
Our results also showed the up-regulated expression of HSP27 in chemotherapy-sensitive group, and Western blotting showed consistent results with the proteomic findings, indicating that proteomic analysis can yield reliable results. Most previous studies (14,15) were focused on the differentially expressed proteins between breast cancer tissue and normal breast tissue. The expression of HSP27 in the breast cancer tissue differs from those in breast cancer cell stains and normal breast tissue. The breast cancer tissue may be suppressed by paclitaxel, and is associated with the endogenous HSP27 inside cells, which is consistent with the findings of Tanaka et al. (17). The up-regulated expression of HSP27 in the sensitive group supports the argument that HSP27-positive breast cancer is more sensitive to chemotherapy (16). A recent research (18) indicates that the level of HSP27 expression can affect the metabolism, growth, and drug resistance of cells by affecting the expression of phosphatase and tensin homologue (PTEN) expression in breast cell lines. Of course, due to the limited sample size of this experiment, some errors may exist, and therefore studies with larger sample sizes are warranted to verify our findings.
VIME is mainly expressed in mesodermal cells such as endothelial cells, fibroblasts, and leukocytes (19). Vimentin is a type III intermediate filament (IF) protein that is expressed in various epithelial-derived malignant tumors. Thompson et al. (20) defines the transformation from epithelial cells to interstitial cells as the expression of VIME in epithelial tumor cells and its role in promoting the invasion and metastasis of tumor cells. Up to now the specific mechanism via which the epithelial cells is transformed to interstitial cells has not been fully elucidated. The expression of VIME in breast cancer may be one of the main causes of multi-drug resistance, which also indicates that the differentiation of breast tumor cells is relatively low (21,22). ACTB, an isomeric form of actin, is distributed in human muscle cells and non-muscle cells. Actin is the main protein in the muscle thin filament and the microfilament in eukaryotic cytoskeleton, and also the richest protein in eukaryotic cells. During the atypical hyperplasia of breast, the change of actin expression may be related with the occurrence of breast cancer (23), and the mammary myoepithelial cells may be also involved in this process. However, their roles in tumor drug-resistance have not been reported.
In summary, our study further reveals some mechanisms related with breast cancer chemotherapy resistance or sensitivity, which may provide new research targets for reversing breast cancer resistance (24,25). On the other hand, some chemotherapy sensitivity or resistance-related specific proteins were identified, which may provide clues for screening candidate proteins in search for the diagnostic markers and treatment targets of breast cancer. Nevertheless, this was only a preliminary study. Further investigations on the above 13 differentially expressed proteins and their mechanisms may be of scientific and clinical importance.
Acknowledgements
This work was supported by the Major Project of Natural Science Fundation of Hunan Province (10jj2031) and the Development and Reform Commission of Hunan Province (2009-1390), P.R.China.
Disclosure: The authors declare no conflict of interest.
References
- Lehnert M. Chemotherapy resistance in breast cancer. Anticancer Res 1998;18:2225-6. [PubMed]
- Grim J, Jandík P, Slánská I, et al. Low expression of NQO1 predicts pathological complete response to neoadjuvant chemotherapy in breast cancer patients treated with TAC regimen. Folia Biol (Praha) 2012;58:185-92. [PubMed]
- Wang D, Jensen RH, Williams KE, et al. Differential protein expression in MCF7 breast cancer cells transfected with ErbB2, neomycin resistance and luciferase plus yellow fluorescent protein. Proteomics 2004;4:2175-83. [PubMed]
- Vergara D, Simeone P, Del Boccio P, et al. Comparative proteome profiling of breast tumor cell lines by gel electrophoresis and mass spectrometry reveals an epithelial mesenchymal transition associated protein signature. Mol Biosyst 2013;9:1127-38. [PubMed]
- Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst 2000;92:205-16. [PubMed]
- Luppens SB, ten Cate JM. Effect of biofilm model, mode of growth, and strain on streptococcusmutans protein expression as determined by two-dimensional difference gel electrophoresis. J Proteome Res 2005;4:232-7. [PubMed]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Biotechnology 1992;24:145-9. [PubMed]
- Giometti CS, Tollaksen SL, Chubb C, et al. Analysis of proteins from human breast epithelial cells using two-dimensional gel electrophoresis. Electrophoresis 1995;16:1215-24. [PubMed]
- Wu SL, Hancock WS, Goodrich GG, et al. An approach to the proteomic analysis of a breast cancer cell line (SKBR-3). Proteomics 2003;3:1037-46. [PubMed]
- Zhang JT. Biochemistry and pharmacology of the human multidrug resistance gene product, ABCG2. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2007;32:531-41. [PubMed]
- Zhang DH, Tai LK, Wong LL, et al. Proteomics of breast cancer: enhanced expression of cytokeratin19 in human epidermal growth factor receptor type 2 positive breast tumors. Proteomics 2005;5:1797-805. [PubMed]
- Furukawa T, Yoshimura A, Sumizawa T, et al. Angiogenic factor. Nature 1992;356:668. [PubMed]
- Blanquicett C, Gillespie GY, Nabors LB, et al. Induction of thymidine phosphorylase in both irradiated and shielded, contralateral human U87MG glioma xenografts: implications for a dual modality treatment using capecitabine and irradiation. Mol Cancer Ther 2002;1:1139-45. [PubMed]
- Huot J, Roy G, Lambert H, et al. Increased survival after treatments with anticancer agents of Chinese hamster cells expressing the human Mr 27,000 heat shock protein. Cancer Res 1991;51:5245-52. [PubMed]
- Chung YM, Park S, Park JK, et al. Establishment and characterization of 5-fluorouracil-resistant gastric cancer cells. Cancer Lett 2000;159:95-101. [PubMed]
- Seymour L, Bezwoda WR, Meyer K. Tumor factors predicting for prognosis in metastatic breast cancer. The presence of P24 predicts for response to treatment and duration of survival. Cancer 1990;66:2390-4. [PubMed]
- Tanaka Y, Fujiwara K, Tanaka H, et al. Paclitaxel inhibits expression of heat shock protein 27 in ovarian and uterine cancer cells. Int J Gynecol Cancer 2004;14:616-20. [PubMed]
- Cayado-Gutiérrez N, Moncalero VL, Rosales EM, et al. Downregulation of Hsp27 (HSPB1) in MCF-7 human breast cancer cells induces upregulation of PTEN. Cell Stress Chaperones 2013;18:243-9. [PubMed]
- Coulombe PA, Wong P. Cytoplasmic intermediate filaments revealed as dynamic and multipurpose scaffolds. Nat Cell Biol 2004;6:699-706. [PubMed]
- Thompson EW, Newgreen DF, Tarin D. Carcinoma invasion and metastasis: a role for epithelial-mesenchymal transition? Cancer Res 2005;65:5991-5; discussion 5995. [PubMed]
- Liu G, Ding H, Gao L, et al. Relationship of vimentin, P-gp and various prognostic factors in breast cancer. Chinese Journal of Histochemistry and Cyto-chemistry 2001;10:179-81.
- Huang C, Cao P, Xie Z, et al. Effect of different heating methods combined with neferine on the expressions of γH2AX and mdr-1/P-gp in MCF-7/Adr breast cancer cells. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2011;36:317-22. [PubMed]
- Kawabata K, Sado Y, Nagayama M, et al. Visuation of dynamic organization of cytoskeleton gels in living cells by hybrid-SPM. Chin J Polym Sci 2003;21:169-74.
- Huang J, Zhang Y, Huang Y, et al. Reversal effect of mifepristone on adriamycin resistance in human breast cancer cell line MCF-7/ADM in vitro and in vivo. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2010;35:576-83. [PubMed]
- Chen J, Shen L, Zhou R, et al. Reversal effect and mechanism of lobeline on the multidrug-resistance of human breast cancer cells MCF-7/ADM. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2009;34:738-43. [PubMed]