
Downregulation of microRNA-30c-5p was responsible for cell migration and tumor metastasis via COTL1-mediated microfilament arrangement in breast cancer
Introduction
Breast cancer, cervical cancer, ovarian cancer, and endometrial cancer are the most common cancers in women. The incidence of breast cancer is the highest among these malignancies, especially in developed countries (1). According to Chinese government reports, about 200,000 new cases of breast cancer occur in China each year, and the growing rate of breast cancer in China is twice that of global average level (2,3). The detection and prevention of breast cancer in the early stage are major public health issues (4), and the Chinese government has listed breast cancer in the Breast and Cervical Cancer Screening program. While research has revealed the mechanisms in breast cancer occurrence, development, metastasis, and drug resistance (5), the mechanisms are still largely unknown.
Metastasis is a characteristic of malignant tumors that severely affects the therapy and prognosis of the patients (6). Cell migration is the biological basis of cancer metastasis, and is considered to be regulated by many mechanisms. microRNAs (miRNAs), short single-chain RNAs, are involved in cancer metastasis, including that of breast cancer (7). MiRNAs can bind to the 3’ UTR region of key genes that regulate cell migration and promote or inhibit cell migration. One transcriptional analysis revealed that an unexpectedly large number of miRNAs were abnormally regulated in breast cancer tissues and paraplastic tissues. The upregulated miRNAs included miR-21and miR-155, while the downregulated miRNAs included miR-10b, miR-125b, and miR-145 (8). Moreover, their target genes participated in multiple cell processes related to tumor genesis and development (9,10). miR-30c had already been studied in many tumors and was reported abnormally expressed, including breast cancer, bladder cancer, colorectal cancer and so on, while its’ effects in breast cancer and the underlying mechanisms was not clear. Studies showed that circPRMT5 absorbed miR-30c and promote bladder cancer invasion (11), and miR-30c suppressed colorectal cancer by targeting BCL9 (12). These studies indicated that miR-30c may exert an anti-tumor ability. A recent review put forward the role of miR-30c in cancer progression (13), and some studies also reported that miR-30c was abnormally expressed in breast cancer (14,15). So in this study, we focus on miR-30c and explore its’ role in breast cancer.
Microfilaments are a major component of the cytoskeleton, are responsible for cell migration and invasion regulation, and play crucial roles in cancer cell metastasis. Previous studies have indicated that microfilament polymerization could promote breast cancer cell migration and invasion (16), and that cytoskeleton focal adhesion proteins were candidate indicators of progression and prognosis in human breast cancer (17). These studies indicate strong correlations between cell microfilaments and breast cancer progression and prognosis; however, the manner in which microfilaments regulate cell migration and affect breast cancer progression is not clearly understood. Coactosin-like protein 1 (COTL1), a kind of F-actin binding protein, can interact with F-actin, participate in cytoskeletal movement, and regulate the inflammation reaction of leukocyte chemotaxis and phagocytosis (18). Studies have shown that the enhanced COTL1 expression might foster cancer progression in non-small lung cancer (19) or suppress breast cancer growth (20). However, whether COTL1 is associated with the metastatic potential of breast cancer is unknown.
MCF-7 was the most typical and widely used cell line to study breast cancer in many previous studies. It is a cell line that was established from human breast cancer, and was very suitable for human breast cancer studying. In this study, we chose MCF-7 as a proper model to determine the expression of the COTL1 gene and its predicted miRNA in breast cancer tissues, and revealed their effects on breast cancer metastasis regulation.
Methods
Tissue collection
This study was conducted in accordance with the Declaration of Helsinki and was approved by the Ethical Committee of Changzhou No. 2 People’s Hospital. All patients were informed of the details of the study and signed the informed consent. The breast cancer tissues and the corresponding paraplastic tissues were collected from patients that had undergone surgical operation from January, 2016 to December, 2018. Totally 40 cancer tissues and the corresponding normal tissues were collected. All the tissues were confirmed by pathological examination. We excluded patients who received chemotherapy, radiotherapy, biological therapy, hormone therapy, or targeted drug therapy, along with those patients who had other non-breast cancer tumors. The patients’ general information and the tumor tissues’ pathological classifications were also recorded.
Cell culture
MCF-7 cells were purchased from Cell Bank of Typical Culture Preservation Committee of Chinese Academy of Sciences (Shanghai, China). The cells were cultured with Minimal Essential Medium (MEM) cell culture medium supplied with 10% fetal bovine serum (FBS), in a 5% CO2, 37 °C atmosphere. Cell culture medium was replaced once a day, and cell transfection was carried out at 60% confluence.
Total RNA extraction and reverse transcriptase polymerase chain reaction (RT-PCR) analysis
The total RNA in the tissue was extracted with TRIzol regent. After the tissues were cut into pieces, 1 mL of the regent was added and the suspension was sonicated in an ice bath. After centrifugation at 15,000 g, the supernatant was separated for the following extraction. Nanodrop 2000 was used for the concentration determination. cDNAs of the genes or the microRNAs were reversed by the commercial kits according to the protocol, respectively. The microRNA was reversed by a specific stem-loop primer, and the U6 was reversed by a random primer to form cDNAs. Then, the target gene or microRNA expression levels were analyzed with SYBR PCR Master Mix reagent kit with the corresponding primers, while anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and U6 were used for loading controls. The primers used in the study are listed as follows: COTL1 F: GCATTTCTCTTCCACGCGAC R: GGTACACAGGAAGCAGGGTC; has-microRNA-30c-5p (miR-30c-5p) reverse: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGCTGAGAG F: ACACTCCAGCTGGGTGTAAACATCCTACACT, URP: TGGTGTCGTGGAGTCG; U6: F: CTCGCTTCGGCAGCACA, R: AACGCTTCACGAATTTGCGT. All primers were synthesized by Invitrogen Company (Shanghai, China).
Oligonucleotide, plasmids synthesis and transfection
MiR-30c-5p mimics, inhibitors, and their negative controls were synthesized by Invitrogen (Shanghai, China). For oligonucleotide transfection, cells were cultured to 60% confluence. Next, the miR-30c-5p mimics, inhibitors, and their negative controls were added and cultured. After 24-h incubation, cells were collected, and then were suspended, lysed, or fixed for Transwell assay, RT-PCR, and Western blotting analysis, or immunofluorescence analysis. Firefly luciferase vectors containing the wild-type (WT) or mutant (Mut) 3’UTR sequences of the COTL1 gene and rinilla luciferase vectors were purchased from GenePharma (Shanghai). The transfection was carried out with Lipofectamine 2000 according to the protocol. After cells were cultured to 50% confluence, vectors and the miR-30c-5p mimics were added and cultured for 6 h. Then the medium was replaced and the cells were cultured for another 24 h before dual-luciferase reporter gene analysis.
Bioinformatics prediction and dual-luciferase assay
The expression between normal breast tissues and cancer tissues was also examined from a chip of tumor expression profiles. The microRNA-gene interaction prediction was carried out with the following databases: TargetScan (http://www.targetscan.org), and miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/index.html), and miRbase (http://www.microrna.org/). For the dual luciferase assay, cells were cultured and transfected with plasmids or negative controls with Lipofectamine 2000. After 24-h incubation, cells were collected and lysed with buffer, and the supernatants were collected for analysis with a dual-luciferase reporter kit (Promega, Madison, WI, USA) according to the manufacture’s protocol.
Cell proliferation measurement
Cells were cultured in a 96 multi-well dish for 12 h, miR-30c-5p and the negative control (NC) was added to finish transfection. After the medium was replaced (recorded as 0 h), cell proliferation was measured with CCK8 assay kit every 6 h at the absorbance of 490 nm for another 24 h according to the manufacture’s guideline.
Western blotting analysis
Tissues were washed 3 times with phosphate-buffered saline (PBS) and cut into pieces. The lysis buffer was added, and ultrasonic treatment was conducted. MCF-7 cells were cultured, and received corresponding treatment. Cell pellets were then collected and sonicated. The tissue and cell supernatants were collected, the total protein concentrations were determined by bicinchoninic acid (BCA) assay kit, total proteins (80 µg) were loaded, and the GAPDH was used as the loading control. The primary COTL1 antibody (Abcam, ab235833) was purchased from Abcam Company.
Immunofluorescence analysis
Cells were cultured in glass bottle dishes (In Vitro Scientific) and transfected with miR-30c-5p mimics and inhibitor. After cells were cultured for 24 h, the cells were fixed with 4% paraformaldehyde for 30 min. After cells were treated with Triton X-100, cell microfilaments were stained with rhodamine phalloidin, while cell nuclei were stained with 4', 6-diamidino-2-phenylindole (DAPI).
Transwell assay
Transwell assay was used to evaluate the invasion ability of MCF-7 cells. Cells were cultured and transfected with miR-30c-5p mimics and inhibitor for 24 h, and were collected and suspended with serum-free medium. After cell densities were adjusted, 100 µL cell suspension was added to the upper chamber and 200 µL complete cell culture medium was added to the lower well. After another 12-h culture, cells were fixed and stained with crystal violet, and cells were counted under a light microscope.
Wound healing assay
Wound healing assay was used to examine the migration ability of MCF-7 cells. Cells were cultured in dishes and transfected with microRNA mimics or inhibitors at80% confluence. Then, the disinfected tips were used to scratch the dish to form a wound. The dishes were cultured for another 12 h after the cell culture medium was refreshed. The distance of the edge was observed under a light microscope.
Statistical analysis
All data are presented as mean ± standard deviation (SD) or standard error (SE) as needed. Analyses of the data from cell studies were repeated 3 times. The difference between 2 groups was analyzed with a two-tailed t-test with GraphPad Prism 5, and a P value <0.05 was considered statistically significant. The correlation between miR-30c-5p and COTL1 protein expression was independently performed with other parameters using scatter plots; the correlation coefficient and linear regression analysis was performed using Pearson’s or Spearman’s rank correlation test. The scatter plot image was drawn with Stata 14.0 software.
Results
Subject characteristics
The basic information of the patients was recorded. Briefly, the age of the patients ranged from 35 to 60 years. The average age of the patients was 55.24±8.76. Among all 40 tissues, 6 were well-differentiated, 28 were moderately differentiated, and 6 were poorly differentiated. A total of 8 patients exhibited lymph node metastasis. There were 22 cases of stage I to II, and 18 cases of stage III to IV based on the tumor-node-metastasis (TNM) classification.
The pathological examination of cancer and paracancerous tissues
Figure 1A shows the pathological image of normal breast tissues, while Figure 1B shows the cancer tissue. The cancer tissue lost the original morphology, and presented aggressive actions to the mammary stroma.
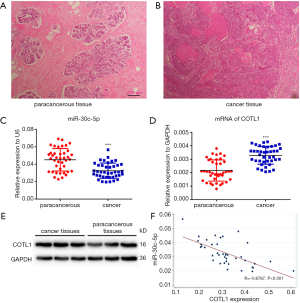
The expression of miR-30c-5p and COTL1 in cancer and paracancerous tissues
The RT-PCR results showed that the expression of miR-30c-5p significantly decreased (P<0.05) in cancer tissues (Figure 1C). We also found that, compared with paracancerous tissues, the mRNA levels of COTL1 significantly increased in tumor tissues (Figure 1D). The protein levels of COTL1 also increased in tumor tissues (Figure 1E). Pearson’s correlation analysis (Figure 1F) showed that miR-30c-5p levels had a significantly negative correlation with COTL1 protein levels (P<0.001).
MiR-30c-5p targeted COTL1
The bioinformatics prediction showed the binding sites of miR-30c-5p on the COTL1 gene (Figure 2A), and the dual-luciferase report assay showed that, compared with mutant group, the average fluorescence intensity decreased in the wild-type group (Figure 2B).
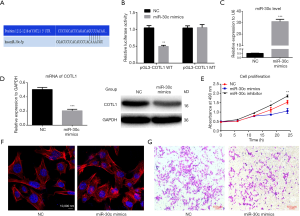
Cell migration and invasion ability after miR-30c-5p was upregulated
After the miR-30c-5p level was upregulated by miR-30c-5p mimics (Figure 2C), the mRNA expression level of COTL1 was decreased (Figure 2D). Further cell proliferation evaluation showed that miR-30c-5p mimics inhibited cell proliferation ability, while knockdown miR-30c-5p promoted cell proliferation (Figure 2E). And after the transfection cellular microfilament arrangement was disordered (Figure 2F), and the cell migration ability was weakened (Figure 2G).
Cell invasion ability after miR-30c-5p was downregulated
After the miR-30c-5p level was downregulated by miR-30c-5p inhibitor (Figure 3A), the expression of COTL1 was increased (Figure 3B). Although cell microfilaments showed no obvious change (data not shown), the cell migration ability was enhanced (Figure 3C).
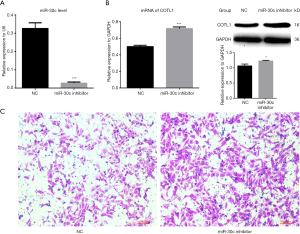
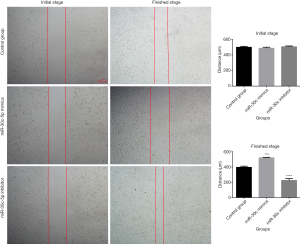
Cell migration ability after miR-30c-5p was changed
The wound healing assay was used to evaluate the cellular migration abilities after miR-30c-5p was modified. The results showed that (Figure 4), after the expression of miR-30c-5p was upregulated by microRNA mimics, the migration ability of MCF-7 cells was decreased, and, when the miR-30c-5p was downregulated by microRNA inhibitor, the cell migration ability was increased.
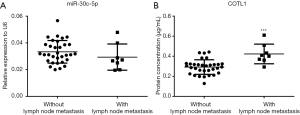
The expression of COTL1 in patients with lymph node metastasis
Figure 5A shows that the expression levels of miR-30c-5p were the same in the cancer tissues with or without lymph node metastasis, while the COTL1 proteins were significantly higher in the cancer tissues with lymph node metastasis (Figure 5B).
Discussion
Breast cancer is a common tumor that severely affects women’s health. This tumor is considered to be a complex result of genetic and environmental factors (21). Studies have indicated that breast cancer exhibits familial aggregation, and women with breast cancer susceptibility gene 1 (BRCA1) or BRCA2 gene mutations are at higher risk (22). In addition, environmental pollutants, especially environmental endocrine disruptors (EDCs) were reported to be associated with breast cancer (23). Despite the abundant research on breast cancer, the mechanisms underlying breast cancer genesis, metastasis, and development of drug resistance are still largely unknown. In this study, we focused on the regulation of cell migration ability and revealed a possible mechanism that regulates cancer metastasis.
We selected the candidate microRNA that was reported to be differently expressed between breast cancer tissue and paracancerous tissue but was not further studied in a tumor specific chip (24). We ultimately chose miR-30c-5p and the targeted COTL1 gene for further study based on an informatics database selection. We examined the expression of both miR-30c-5p and COTL1 in breast cancer and paracancerous tissue, and found that miR-30c-5p was downregulated while COTL1 was increased in cancer tissues. MiR-30c-5p is reported to be abnormally expressed in gastric cancer (25), breast cancer (26), and prostatic cancer (27). It can regulate the downstream cancer-related genes and has been demonstrated to participate in tumor development, metastasis, and drug resistance. Tanic and his colleges compared the microRNA expression profiles in normal breast tissues and cancer tissues, and found 19 differently expressed miRNAs, including miR-30c-5p (28). Meanwhile, other research has indicated that miR-30c-5p was downregulated in many other tumors, and a further study showed that miR-30c-5p participated in the cell migration regulation and was associated with patients’ prognosis (25). However, it is still unknown whether miR-30c-5p regulates cell migration progress in breast cancer.
Metastasis is a feature of malignant tumor and is biologically based in cell migration. Xia et al. carried out an experiment in A549 cells and found that a low expression of miR-30c-5p was associated with highly aggressive non-small cell lung carcinoma (NSCLC) (29). Moreover, other researchers found that the overexpression of miR-30c-5p in endometrial cancer could significantly inhibit cell migration and invasion ability (30). According to Bockhorn, miR-30c-5p could be a biomarker for prognosis prediction, as miR-30c-5p expression was shown to be related to neoplasm staging (31). Despite these findings, the process that regulates breast cancer cells remains largely unknown.
MiR-30c-5p has been shown to regulate many tumor-related pathways including KRAS, MTA, p38/MAPK, HER, and RAC1 (28,30,32,33). In this study, we screened the COTL1 gene by informatics prediction, and we found that miR-30c-5p could bind to the 3’ UTR region of COTL1 and regulate both the mRNA and protein levels of COTL1. COTL1, which is encoded by the COTL1 gene, is a kind of F-actin binding protein, and mainly regulates cell motivation and migration (34). Microfilaments compose the main cytoskeletal component that regulates cell migration. Researchers have found that luteinizing hormone (LH) and follicle-stimulating hormone (FSH) could promote breast cancer cell migration via the microfilament-related pathways (35). Microfilaments have even been found to regulate pre-invasive breast cancer cell stiffening and proliferation via the actin organization (36). Apart from the microfilament, many cytoskeleton-related genes, including TWF1 and VIM were reported to be involved in tumor migration ability and cancer progression regulation (37,38). COTL1 is also a microfilament-related protein and was reported to be highly expressed in lung cancer (34), but its role in breast cancer is still largely unknown. In this study, we found that after the miR-30c-5p/COTL1 pathway was reversed in 2 directions, cell migration was changed accordingly. As microRNAs are currently being designed for disease treatment (39), the miR-30c-5p could also be used as a target to hamper breast cancer metastasis
While our study did yield many promising insights a main limitation should be addressed. Chiefly, we did not obtain the follow-up data of the patients that we recruited into this study when we reported the current results, so the current results was a cross-sectional view of the patients with cancer metastasis, and we could not obtain solid evidence to prove the cause and effect relation between microRNA/gene expression and disease prognosis.
Conclusions
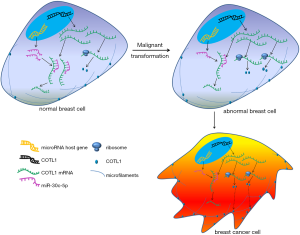
Our study indicates that miR-30c-5p/COTL1 may be crucially involved in the regulation of breast cancer cell migration ability. In breast cancer cells, the expression of miR-30c-5p was downregulated, the inhibition of miR-20c-5p for COTL1 was weakened, and COTL1 protein level was then increased, which enhanced cell migration ability and finally induced tumor metastasis (Figure 6). Our study offers new insights into breast cancer metastasis regulation and may provide potential targets for future treatment.
Acknowledgments
Funding: None.
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/gs-20-472
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/gs-20-472). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki and was approved by the Ethical Committee of Changzhou No. 2 People’s Hospital. All patients were informed of the details of the study and signed the informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Naito Y, Urasaki T. Precision medicine in breast cancer. Chin Clin Oncol 2018;7:29. [Crossref] [PubMed]
- Renzulli M, Zanotti S, Clemente A, et al. Hereditary breast cancer: screening and risk reducing surgery. Gland Surg 2019;8:S142-9. [Crossref] [PubMed]
- Ganz PA, Goodwin PJ. Breast Cancer Survivorship: Where Are We Today? Adv Exp Med Biol 2015;862:1-8. [Crossref] [PubMed]
- Merino Bonilla JA, Torres Tabanera M, Ros Mendoza LH. Breast cancer in the 21st century: from early detection to new therapies. Radiologia 2017;59:368-79. [Crossref] [PubMed]
- de Boer MC, Worner EA, Verlaan D, et al. The Mechanisms and Effects of Physical Activity on Breast Cancer. Clin Breast Cancer 2017;17:272-8. [Crossref] [PubMed]
- Eiro N, Gonzalez LO, Fraile M, et al. Breast Cancer Tumor Stroma: Cellular Components, Phenotypic Heterogeneity, Intercellular Communication, Prognostic Implications and Therapeutic Opportunities. Cancers (Basel) 2019.11. [PubMed]
- Du Y, Zhou L, Lin Y, et al. Polymorphisms in microRNA let-7 binding sites of the HIF1AN and CLDN12 genes can predict pathologic complete response to taxane- and platinum-based neoadjuvant chemotherapy in breast cancer. Ann Transl Med 2019;7:138. [Crossref] [PubMed]
- O'Day E, Lal A. MicroRNAs and their target gene networks in breast cancer. Breast Cancer Res 2010;12:201. [Crossref] [PubMed]
- Petrović N. miR-21 Might be Involved in Breast Cancer Promotion and Invasion Rather than in Initial Events of Breast Cancer Development. Mol Diagn Ther 2016;20:97-110. [Crossref] [PubMed]
- Zaleska K, Przybyla A, Kulcenty K, et al. Wound fluids affect miR-21, miR-155 and miR-221 expression in breast cancer cell lines, and this effect is partially abrogated by intraoperative radiation therapy treatment. Oncol Lett 2017;14:4029-36. [Crossref] [PubMed]
- Chen X, Chen RX, Wei WS, et al. PRMT5 Circular RNA Promotes Metastasis of Urothelial Carcinoma of the Bladder through Sponging miR-30c to Induce Epithelial-Mesenchymal Transition. Clin Cancer Res 2018;24:6319-30. [Crossref] [PubMed]
- Zhao DW, Li MM, Han JP, et al. MiR-30c exerts tumor suppressive functions in colorectal carcinoma by directly targeting BCL9. Eur Rev Med Pharmacol Sci 2019;23:3335-43. [PubMed]
- Han W, Cui H, Liang J, et al. Role of MicroRNA-30c in cancer progression. J Cancer 2020;11:2593-601. [Crossref] [PubMed]
- Bertoli G, Cava C, Castiglioni I. MicroRNAs: New Biomarkers for Diagnosis, Prognosis, Therapy Prediction and Therapeutic Tools for Breast Cancer. Theranostics 2015;5:1122-43. [Crossref] [PubMed]
- Croset M, Pantano F, Kan CWS, et al. miRNA-30 Family Members Inhibit Breast Cancer Invasion, Osteomimicry, and Bone Destruction by Directly Targeting Multiple Bone Metastasis-Associated Genes. Cancer Res 2018;78:5259-73. [Crossref] [PubMed]
- Lang L, Hou Y, Chen Y, et al. ATM-Mediated Phosphorylation of Cortactin Involved in Actin Polymerization Promotes Breast Cancer Cells Migration and Invasion. Cell Physiol Biochem 2018;51:2972-88. [Crossref] [PubMed]
- Omran OM, Al Sheeha M. Cytoskeletal Focal Adhesion Proteins Fascin-1 and Paxillin Are Predictors of Malignant Progression and Poor Prognosis in Human Breast Cancer. J Environ Pathol Toxicol Oncol 2015;34:201-12. [Crossref] [PubMed]
- Li G, Yin Y, Chen J, et al. Coactosin-like protein 1 inhibits neuronal migration during mouse corticogenesis. J Vet Sci 2018;19:21-6. [Crossref] [PubMed]
- Sun W, Guo C, Meng X, et al. Differential expression of PAI-RBP1, C1orf142, and COTL1 in non-small cell lung cancer cell lines with different tumor metastatic potential. J Investig Med 2012;60:689-94. [Crossref] [PubMed]
- Xia L, Xiao X, Liu WL, et al. Coactosin-like protein CLP/Cotl1 suppresses breast cancer growth through activation of IL-24/PERP and inhibition of non-canonical TGFbeta signaling. Oncogene 2018;37:323-31. [Crossref] [PubMed]
- Rojas K, Stuckey A. Breast Cancer Epidemiology and Risk Factors. Clin Obstet Gynecol 2016;59:651-72. [Crossref] [PubMed]
- De Silva S, Tennekoon KH, Karunanayake EH, et al. Analysis of BRCA1and BRCA2 large genomic rearrangements in Sri Lankan familial breast cancer patients and at risk individuals. BMC Res Notes 2014;7:344. [Crossref] [PubMed]
- Rachoń D. Endocrine disrupting chemicals (EDCs) and female cancer: Informing the patients. Rev Endocr Metab Disord 2015;16:359-64. [Crossref] [PubMed]
- Turashvili G, Lightbody ED, Tyryshkin K, et al. Novel prognostic and predictive microRNA targets for triple-negative breast cancer. FASEB J 2018.fj201800120R. [PubMed]
- Cao JM, Li GZ, Han M, et al. MiR-30c-5p suppresses migration, invasion and epithelial to mesenchymal transition of gastric cancer via targeting MTA1. Biomed Pharmacother 2017;93:554-60. [Crossref] [PubMed]
- Yen MC, Shih YC, Hsu YL, et al. Isolinderalactone enhances the inhibition of SOCS3 on STAT3 activity by decreasing miR-30c in breast cancer. Oncol Rep 2016;35:1356-64. [Crossref] [PubMed]
- Cochetti G, Poli G, Guelfi G, et al. Different levels of serum microRNAs in prostate cancer and benign prostatic hyperplasia: evaluation of potential diagnostic and prognostic role. Onco Targets Ther 2016;9:7545-53. [Crossref] [PubMed]
- Tanic M, Yanowsky K, Rodriguez-Antona C, et al. Deregulated miRNAs in hereditary breast cancer revealed a role for miR-30c in regulating KRAS oncogene. PLoS One 2012;7:e38847. [Crossref] [PubMed]
- Xia Y, Chen Q, Zhong Z, et al. Down-regulation of miR-30c promotes the invasion of non-small cell lung cancer by targeting MTA1. Cell Physiol Biochem 2013;32:476-85. [Crossref] [PubMed]
- Zhou H, Xu X, Xun Q, et al. microRNA-30c negatively regulates endometrial cancer cells by targeting metastasis-associated gene-1. Oncol Rep 2012;27:807-12. [PubMed]
- Bockhorn J, Dalton R, Nwachukwu C, et al. MicroRNA-30c inhibits human breast tumour chemotherapy resistance by regulating TWF1 and IL-11. Nat Commun 2013;4:1393. [Crossref] [PubMed]
- Fang Y, Shen H, Cao Y, et al. Involvement of miR-30c in resistance to doxorubicin by regulating YWHAZ in breast cancer cells. Braz J Med Biol Res 2014;47:60-9. [Crossref] [PubMed]
- Rodríguez-González FG, Sieuwerts AM, Smid M, et al. MicroRNA-30c expression level is an independent predictor of clinical benefit of endocrine therapy in advanced estrogen receptor positive breast cancer. Breast Cancer Res Treat 2011;127:43-51. [Crossref] [PubMed]
- Guo S, Yang P, Jiang X, et al. Genetic and epigenetic silencing of mircoRNA-506-3p enhances COTL1 oncogene expression to foster non-small lung cancer progression. Oncotarget 2017;8:644-57. [Crossref] [PubMed]
- Sanchez AM, Flamini MI, Russo E, et al. LH and FSH promote migration and invasion properties of a breast cancer cell line through regulatory actions on the actin cytoskeleton. Mol Cell Endocrinol 2016;437:22-34. [Crossref] [PubMed]
- Tavares S, Vieira AF, Taubenberger AV, et al. Actin stress fiber organization promotes cell stiffening and proliferation of pre-invasive breast cancer cells. Nat Commun 2017;8:15237. [Crossref] [PubMed]
- Bockhorn J, Yee K, Chang YF, et al. MicroRNA-30c targets cytoskeleton genes involved in breast cancer cell invasion. Breast Cancer Res Treat 2013;137:373-82. [Crossref] [PubMed]
- Clucas J, Valderrama F. ERM proteins in cancer progression. J Cell Sci 2014;127:267-75. [Crossref] [PubMed]
- Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017;16:203-22. [Crossref] [PubMed]


