
Von Hippel-Lindau “Black Forest” mutation inherited in a large Chinese family
Introduction
Von Hippel-Lindau (VHL) disease, an autosomal dominant inherited neoplastic syndrome, presents with various clinical phenotypes and age-dependent penetrance. Many kinds of tumors are related to VHL, such as central nervous system (CNS) haemangioblastomas, retinal angiomas (RAs), clear-cell renal cell carcinomas (ccRCCs), pheochromocytomas (PCCs), pancreatic neuroendocrine tumors, pancreatic cysts, and endolymphatic sac tumors (1-3). Meanwhile, paragangliomas (PGLs), and epididymal and broad ligament cystadenomas may also be possible manifestations of VHL disease (4-6). The incidence of VHL disease is approximately 1/36,000 (7). VHL has over 90% penetrance by the age of 65, and the onset of symptoms can even occur during childhood (8).
VHL disease has traditionally been classified into 2 types based on its clinical manifestations (1,9): type 1 (reduced risk of PCCs) and type 2 (with PCCs). Type 2 is further subdivided in to type 2A (low risk of ccRCCs), type 2B (high risk of ccRCCs), and type 2C (only with PCCs, without ccRCCs or haemangioblastomas) (9,10). Patients were diagnosed as having VHL disease according to their clinical characteristics before the genetic testing was complemented in clinics. However, there were several patients who had a clinical diagnosis of VHL disease but lacked VHL mutations, indicating that genetic testing is the gold standard rather than exclusively relying on diagnosis based on clinical features (11). As a result, genetic testing can be used to verify clinical diagnoses in affected VHL patients and rule out VHL disease in individuals with VHL-related tumors without family history (12). After the mutation of VHL has been confirmed in the proband of the family, genetic testing can also be utilized for diagnosis or pre-symptomatic screening of other family members. In addition, genetic testing has shown great potential for use in prenatal screening (12,13).
The VHL p.Tyr98His (Y98H) mutation was designated as the “Black Forest” founder mutation (14), which was previously identified in two VHL type 2A families that lived in Pennsylvania (USA), but was later shown to have originated and spread in the isolated mountain valleys of the Black Forest in Germany (14). Additionally, VHL Y98H was also detected in Sweden and some other non-Asian countries (4,6,15,16).
In Xiangya Hospital, South Central China, we carried out paired whole-exome sequencing (WES) and Sanger sequencing on a Chinese PCC family, which was eventually identified as being a VHL type 2 family with the VHL Y98H mutation. As far as we know, this is the first VHL family with the VHL Y98H mutation that has been reported in Asia. Furthermore, we summarized and analyzed its clinical manifestations and compared it with VHL Y98N mutation.
Methods
Subjects and samples
A large PCC family with four generations was followed by the Department of Urology, Xiangya Hospital, Central South University in China. This study was approved and supervised by the Ethics Committee of Xiangya Hospital (No. 201403307), and informed written consents were signed by each participant. Approximately 2 mL of peripheral venous blood was collected from each family member in tubes that contained anticoagulant (EDTA-K2). DNA samples from peripheral venous blood of all the members were extracted with the Dzup kit (Dzup, Sangon Biotech Co. Ltd, Shanghai, China). The DNA samples were assessed for quantity and quality before sequencing.
Paired WES
Paired WES was first performed on genomic DNA from peripheral venous blood of the proband’s mother and younger brother at the Beijing Genome Institute in Shenzhen, China. The appropriately qualified genomic DNA samples were randomly fragmented by the Covaris Acoustic System, and the library fragments were mainly distributed between 200 and 300 bp. Adapters were then ligated to both ends of the resulting fragments. The adapter-ligated templates were purified by using Agencourt AMPure SPRI beads, and the fragments with insert size of approximately 176 bp were excised. Extracted DNA was amplified by ligation-mediated PCR (LM-PCR), purified, and hybridized to the Nimblegen SeqCap EZ Library v3.0 (Roche/NimbleGen, Madison, WI) for enrichment, and the magnitude of enrichment was estimated for the products. Each qualified library was then loaded on the Hiseq2500 platform (Illumina, San Diego, CA, USA) and sequenced. Finally, the raw image files were processed by Illumina base-calling Software v1.7, and paired-end readings were generated and stored in FASTQ format (raw data).
Bioinformatics analysis
Clean data were produced by filtering the raw data. The Burrows-Wheeler Aligner was used to compare the clean data of each sample to the human reference genome (GRCh37/HG19) (17,18). Local realignment around insertion/deletions (InDels) and base quality score recalibration was performed using the Genome Analysis Toolkit (GATK) (v3.3.0) (19,20). All genomic variations, including single nucleotide polymorphisms (SNPs) and InDels, were detected by the state-of-the-art GATK (v3.3.0) haplotype caller module. Moreover, exome copy number variants (CNVs) were identified by ExomeCNV. Annotation of the genetic variants was performed using the Human Gene Mutation Database (HGMD), dbSNP, ExAC, ClinVar and 1000 Genomes Project databases.
Sanger sequencing
When the causative mutation was confirmed, Sanger sequencing was further performed on all the other family members. The target gene was amplified via polymerase chain reaction (PCR) with the following primers: F-5'-GTACGGCCCTGAAGAAGACG-3'; R-5'-GTCACCCTGGATGTGTCCTG-3'. PCR was performed in a total volume of 20 µL, including 0.3 µL DNA (15 ng), 0.3 µL forward primer (20 µmol/L), 0.3 µL reverse primer (20 µmol/L), 10 µL PrimeSTAR (Takara Biotechnology Co, Ltd., Japan) and 9.1 µL deionized water. The PCR programme was as follows: 98 °C for 5 min; 98 °C for 10 s, 58 °C for 5 s and 72 °C for 1 min for a total of 40 cycles; 72 °C for 5 min. After that, the PCR products were purified and sent to Biosune, Shanghai, China for Sanger sequencing.
Immunohistochemistry (IHC)
The antibodies against SDHB, mouse monoclonal clone 21A11 (NB600-1366; Novus Biologicals, Littleton, CO, USA; 1:50), were applied on the four patients’ formalin fixed paraffin-embedded (FFPE) tissues. At first, the sections were deparaffinised, rehydrated, and heated in a microwave oven in Tris-EDTA buffer (10 mM Tris, 1mM EDTA) at 100 °C for 15 min. Later, after rinsing in tap water, they were incubated in 3% H2O2 in PBS for 20 min. After that, the primary SDHB antibodies were diluted, and slides were incubated with 100 µL per slide at 37 °C for 1 hour, followed by rinsing in 0.5% Tris-Tween buffer (pH =8.0). The sections were then incubated for 45 minutes with peroxidase-labeled polymers coupled with goat anti-mouse immunoglobulin (EnVision Dual Link System-HRP Kit; Dako Cytomation, Carpinteria, CA, USA). Finally, diaminobenzene tetrahydrochloride was applied twice (5 min each time) and the glass slides were rinsed with distilled water.
Results
Presentation of the extended family
The proband was a 34-year-old female with bilateral adrenal masses detected by ultrasonography. The patient did not have headaches, palpitations, predilection to sweating, nor other complaints. Physical examination was normal for her, including blood pressure. Family history of diseases was initially denied. Plasma cortisol, renin, angiotensin and aldosterone were all within normal values, except for urinary vanilmandelic acid (VMA), which was 114.2 µmol/day (normal range: 10–30 µmol/day). Computed tomography of the adrenal glands showed: (I) bilateral adrenal masses, both suggestive of PCCs; (II) a hepatic haemangioma in the left lobe (Figure 1A). Therefore, PCCs were highly suspected. The patient was given Prazosin regularly for 2 weeks, and laparoscopic bilateral partial adrenalectomy (LBPA) was finally performed. Later, histopathological examination confirmed that she had bilateral PCCs.
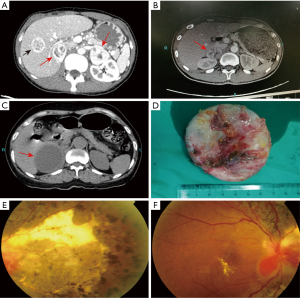
In addition, the proband’s younger brother, a 33-year-old male, was also found to have a right adrenal mass by an imaging examination 7 months after his sister’s LBPA. He was asymptomatic, and the results of physical examination, including blood pressure, were unremarkable. Urinary VMA was 55.0 µmol/day. Computed tomography of the adrenal glands indicated (Figure 1B) that a right PCC was strongly suspected in light of his sister’s condition. Therefore, laparoscopic right partial adrenalectomy (LRPA) was performed after treatment with Prazosin for one month. The diagnosis was confirmed by histopathological examination.
After diagnosis of two PCC patients in this family, we considered the possibility of hereditary pheochromocytoma (HPCC). To test this hypothesis, the patients’ other family members were enrolled and screened by adrenal ultrasonography. As expected, more PCC patients were found in this family. At first, their mother, a 61-year-old female who had been treated for hypertension but lacked any other relevant PCC symptoms, was found to have a large right adrenal mass. After preoperative computed tomography of adrenal glands (Figure 1C) and Prazosin treatment for 1 month, she also had LRPA, and a right cystic tumour was found during the operation (Figure 1D). The right PCC was identified by the histopathological examination after LRPA. Later, it was discovered that she had RA approximately 10 years ago, and a recent fundus examination supported this finding (Figure 1E,F).
In addition, the proband’s cousin, a 31-year-old male, who had LRPA 10 years ago for right PCC, was found to have a left adrenal mass during the imaging examination. However, he had neither clinical symptoms nor any other physical findings associated with PCC in the examinations. In addition, his urinary VMA level within 24 hours was normal. Finally, he underwent a laparoscopic left partial adrenalectomy, and left PCC was confirmed by histopathology.
WES and Sanger sequencing results
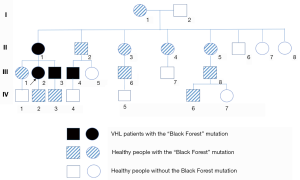
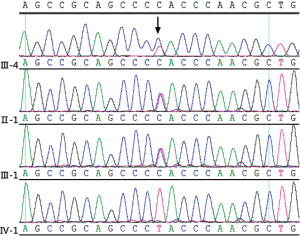
To identify the inherited germline mutation in this family, the genomes of II-1 and III-3 (Figure 2) were sequenced by paired WES. Filtering steps were performed as follows. First, we removed genomic variants that didn’t belong to PCC/PGL susceptibility genes, including RET, VHL, NF1, SDHA, SDHB, SDHC, SDHD, SDHAF2, TMEM127, MAX, FH, EGLN1, MEN1, GDNF, GNAS, CDKN2A, P53, BAP1, BRCA1, BRCA2, ATRX, KMT2D, RAS, EPAS1, MDH2, IDH and KIF1B (18). After this filtering step, there were 136 and 28 SNP and InDel, respectively, variants identified for II-1; the corresponding figures for III-3 were 134 and 32 (Table 1). Then, 5’-prime-UTR, 3’-prime-UTR, intronic and synonymous variants were excluded, remaining 12 SNP and 1 InDel variants for both II-1 and III-3 (Table S1). Next, we used a stringent filtering algorithm, with which we removed variants with minor allele frequency (MAF) >0.1% in 1,000 genomes and ESP6500 databases. Finally, there were 2 genomic variants left, one in VHL and the other in KMT2D, in which were found in both of II-1 and III-5. The VHL NM_000551.3: c.292 T > C mutation was demonstrated as a pathologic mutation, whereas the KMT2D NM_003482.3: c.12911 C > T mutation was found in healthy individuals in the ExAC database (http://exac.broadinstitute.org/variant/12-49425577-G-A), and was not suggested to be a pathologic germline mutation of PCC.

Full table
Full table
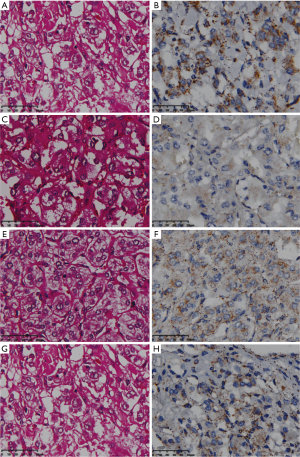
To further exclude interference from other pathogenic genes of PCC/PGL, we performed copy number analysis and discovered that there were no copy number variants identified in genes related to PCC/PGL. Moreover, SDHB IHC was positive in the four patients of this family, ruling out the possibility of SDHB mutation again (Figure 3).
Eventually, Sanger sequencing revealed that in addition to the proband and his mother, 13 other family members had carried the VHL Y98H mutation, of which 2 members were PCC and 11 members remained healthy at the end of this study (Figure 2).
Genotype-phenotype associations
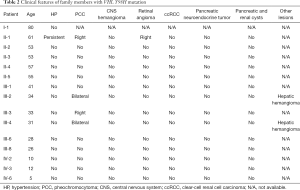
In the present family, 15 members (54%) from 4 generations were identified to have the VHL Y98H mutation, but only 4 members were diagnosed with PCC (Figures 2,4). Most of the mutation-positive individuals have had VHL-related check-ups subsequently, and clinical presentations of the mutation-positive individuals are described (Table 2).
Full table
CNS haemangioblastomas are cardinal features of VHL disease and occur in 60–80% of VHL patients (21,22). Therefore, the medical history of each mutation carrier was obtained again. Initially, II-1 recalled that she had sudden blindness in her right eye approximately 10 years ago, and a RA, which has an identical histopathological appearance to a CNS haemangioblastomas, was eventually diagnosed; however, her other examination results were negative.
Additionally, III-2 was the first diagnosed PCC patient. The lesion in her liver was shown to be a haemangioma by ultrasound-guided percutaneous biopsy. Several rounds of radiofrequency ablation were performed on the benign lesion. Related check-ups were also performed on III-2, III-3 and III-4, and CNS magnetic resonance, fundus examination and abdominal ultrasonography results were negative except for a hepatic hemangioma that was also found on III-4 (Table 2).
The oldest member with VHL Y98H mutation, I-1, was 80 years old. She refused to do any check-ups because she felt she was healthy and could not accept any potentially bad results from the check-ups. The II-2, II-3, II-4, II-5, III-1, III-6 and III-8 individuals were healthy adults with VHL Y98H mutations. They felt healthy, and several examinations were conducted that showed no presence of VHL-related symptoms.
IV-2, IV-3 and IV-6 individuals were 10, 12 and 5 years old, respectively. All of them were healthy individuals with VHL Y98H mutations because they were too young to form any VHL-related tumors.
Overall, among all the family members with VHL Y98H mutation, II-1 was a VHL type 2A patient; III-2, III-3 and III-4 were VHL type 2C patients; and the other eleven individuals were asymptomatic.
Discussion
In this study, the VHL “Black Forest” mutation that was believed to have evolved in the white population was identified in a large Chinese family for the first time. Furthermore, clinical features and genotype-phenotype associations of patients with VHL Y98H mutation were summarized and analysed.
VHL disease is an autosomal dominant inherited neoplastic syndrome caused by inactivation of the VHL tumor suppressor gene mapped to chromosome 3p25.3 (2,23). The clinical diagnosis of VHL disease is traditionally described as follows: (I) at least two haemangioblastomas in the CNS or retina; (II) one haemangioblastoma in the CNS or retina plus at least one VHL-related visceral tumor (excluding epididymal and renal cysts); (III) any one of above items with a family history of VHL (24). Apart from the diseases mentioned above, other VHL-related visceral tumours include endolymphatic sac tumours occurring from head and neck, pancreatic cysts, serous cystadenomas, pancreatic neuroendocrine tumours (NETs), renal cysts, ccRCCs, epididymal cysts, papillary cystadenomas of the epididymis and broad ligament cystadenomas, indicating the variability and complexity of VHL disease (25). Therefore, when diagnosing VHL disease, a systematic examination should be done to avoid a missed diagnosis of VHL-related visceral tumors. In addition, the occurrence of multiple diseases is more conducive to the diagnosis.
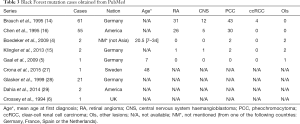
Different mutations cause different types of VHL disease. Patients with truncating mutations, exon deletions, or missense mutations that disrupt the integrity of the VHL protein structure probably present with type 1 VHL disease, but individuals with type 2 VHL disease would carry the surface missense mutations (mutation site at the surface of the VHL protein) (24-26). VHL Y98H is a surface missense mutation, whose carriers have been widely regarded as type 2A. The VHL Y98H mutation was also called the “Black Forest” founder mutation (14). In the past, this mutation was reported only in the Occident (Table 3). However, the present study is the first report of a large Chinese family with VHL Y98H mutations, proving that there is no geographical limitation for this mutation. In fact, approximately 20% of patients have VHL disease as the result of a de novo mutation, which means de novo mutation at codon 98 can occur independently of the race or of the geographic location. Therefore, the occurrence of this mutation is likely to be random. Furthermore, there are no distinct differences in clinical manifestations for the present patients compared with former cases (Table 3). However, there are still a small number of patients with Y98H mutations having ccRCCs, indicating that the genotype-phenotype mismatch of VHL Y98H mutations is not impossible. In addition, the similar mutation in the same codon, VHL Y98N mutation, is associated with type 2B (high risk of ccRCC). Knauth et al. found that the Y98H mutation results in higher binding affinities for key cellular substrates when compared to the Y98N mutation, indicating the internal mechanism of the difference between the two types of manifestations (30,31).
Full table
It is recommended that all VHL mutation carriers should be offered surveillance in which regular follow-up visits were adopted to check each organ involved in the VHL disease. To be specific, (I) ophthalmoscopy is recommended to be done yearly from infancy; (II) plasma or urinary catecholamines and metanephrines should be done (yearly and when blood pressure is raised) from 2 years of age; (III) MRI of craniospinal axis is recommended to be done yearly from 11 years of age; (IV) ultrasound of abdomen is advised to be done yearly from 8 years of age; and (V) CT of abdomen should be done yearly from 18 years of age or earlier if it is clinically indicated (13). As it is widely documented that many mutations are related to its clinical types, surveillance could also be done according to the genotype of VHL patients, suggesting that genetic consulting is imperative for VHL patients. As a result, personalized surveillance has been recommended to the family members with VHL Y98H, especially for the younger ones (IV-2, IV-3 and IV-6).
In the present study, WES and Sanger sequencing were utilized to diagnose VHL disease, though they are not often used to diagnose VHL disease in China. We were initially unable to confirm presence of a pathogenic gene of the PCC family. Therefore, WES was performed because it was more efficient than Sanger sequencing to screen all susceptible genes. Moreover, WES of two or more familial patients could exclude non-shared mutations to narrow the scope of candidate genes. In addition, combination of WES and Sanger sequencing offers the opportunity to detect pathogenic mutations in familial and sporadic PCC cases, especially for clinically atypical or asymptomatic individuals. Meanwhile, with the achievements of genotype-phenotype studies, precision medicine of PCC and VHL was estimated to have a bright future, such as treating tumours in a timely fashion.
In conclusion, we report the first recorded Chinese VHL type 2 family with the “Black Forest” mutation using WES and Sanger sequencing, thus widening the currently recorded presence of the “Black Forest” mutation to China and potentially elsewhere in Asia and indicating that the “Black Forest” mutation does not uniquely evolve in occident countries. In addition, a personalized surveillance approach, which may be more appropriate for affected families, has been recommended to improve quality of life.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 81400773), the Natural Science Foundation of China Hunan Province (No. 14JJ6002), the Science and Technology Plan Project of Hunan Province, China (No. 2017SK2092), funds for the Shenghua Yuying talents program of Central South University and the Fundamental Research Funds for the Graduate Students of Central South University (No. 2017zzts901).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved and supervised by the Ethics Committee of Xiangya Hospital (No. 201403307), and informed written consents were signed by each participant.
References
- Jia D, Tang B, Shi Y, et al. A deletion mutation of the VHL gene associated with a patient with sporadic von Hippel-Lindau disease. J Clin Neurosci 2013;20:842-7. [Crossref] [PubMed]
- Yuan P, Sun Q, Liang H, et al. Germline mutations in the VHL gene associated with 3 different renal lesions in a Chinese von Hippel-Lindau disease family. Cancer Biol Ther 2016;17:599-603. [Crossref] [PubMed]
- Chen Y, Liu H, Zhang K, et al. Massive exudative retinal detachment following photodynamic therapy for retinal hemangioma in von Hippel-Lindau Syndrome. Photodiagnosis Photodyn Ther 2014;11:250-3. [Crossref] [PubMed]
- Boedeker CC, Erlic Z, Richard S, et al. Head and neck paragangliomas in von Hippel-Lindau disease and multiple endocrine neoplasia type 2. J Clin Endocrinol Metab 2009;94:1938-44. [Crossref] [PubMed]
- Gaal J, van Nederveen FH, Erlic Z, et al. Parasympathetic paragangliomas are part of the Von Hippel-Lindau syndrome. J Clin Endocrinol Metab 2009;94:4367-71. [Crossref] [PubMed]
- He Z, Xia L, Deng Z, et al. Identification of a VHL gene mutation in a Chinese family with Von HippelLindau syndrome. Mol Med Rep 2018;18:435-40. [PubMed]
- Maher ER, Webster AR, Moore AT. Clinical features and molecular genetics of Von Hippel-Lindau disease. Ophthalmic Genet 1995;16:79-84. [Crossref] [PubMed]
- Lonser RR, Glenn GM, Walther M, et al. von Hippel-Lindau disease. Lancet 2003;361:2059-67. [Crossref] [PubMed]
- Neumann HP, Bausch B, McWhinney SR, et al. Germ-line mutations in nonsyndromic pheochromocytoma. N Engl J Med 2002;346:1459-66. [Crossref] [PubMed]
- Liu P, Li M, Guan X, et al. Clinical Syndromes and Genetic Screening Strategies of Pheochromocytoma and Paraganglioma. J Kidney Cancer VHL 2018;5:14-22. [Crossref] [PubMed]
- Hes FJ, van der Luijt RB, Janssen AL, et al. Frequency of Von Hippel-Lindau germline mutations in classic and non-classic Von Hippel-Lindau disease identified by DNA sequencing, Southern blot analysis and multiplex ligation-dependent probe amplification. Clin Genet 2007;72:122-9. [Crossref] [PubMed]
- Liu L, Chen C, Li Y, et al. Whole-Exome Sequencing Identified a De Novo Mutation of Junction Plakoglobin (p.R577C) in a Chinese Patient with Arrhythmogenic Right Ventricular Cardiomyopathy. Biomed Res Int 2019;2019:9103860. [Crossref] [PubMed]
- Zhang W, Li D, Wei S, et al. Correction: Whole-exome sequencing identifies a novel CCDC151 mutation, c.325GT (p.E109X), in a patient with primary ciliary dyskinesia and situs inversus. J Hum Genet 2019;64:829. [Crossref] [PubMed]
- Brauch H, Kishida T, Glavac D, et al. Von Hippel-Lindau (VHL) disease with pheochromocytoma in the Black Forest region of Germany: evidence for a founder effect. Hum Genet 1995;95:551-6. [Crossref] [PubMed]
- Klingler JH, Kruger MT, Lemke JR, et al. Sequence variations in the von Hippel-Lindau tumor suppressor gene in patients with intracranial aneurysms. J Stroke Cerebrovasc Dis 2013;22:437-43. [Crossref] [PubMed]
- Chen F, Kishida T, Yao M, et al. Germline mutations in the von Hippel-Lindau disease tumor suppressor gene: correlations with phenotype. Hum Mutat 1995;5:66-75. [Crossref] [PubMed]
- Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol 2011;29:24-6. [Crossref] [PubMed]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 2013;14:178-92. [Crossref] [PubMed]
- DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491-8. [Crossref] [PubMed]
- McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297-303. [Crossref] [PubMed]
- Richard S, David P, Marsot-Dupuch K, et al. Central nervous system hemangioblastomas, endolymphatic sac tumors, and von Hippel-Lindau disease. Neurosurg Rev 2000;23:1-22; discussion 23-4. [Crossref] [PubMed]
- Wanebo JE, Lonser RR, Glenn GM, et al. The natural history of hemangioblastomas of the central nervous system in patients with von Hippel-Lindau disease. J Neurosurg 2003;98:82-94. [Crossref] [PubMed]
- Latif F, Tory K, Gnarra J, et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science 1993;260:1317-20. [Crossref] [PubMed]
- Nordstrom-O'Brien M, van der Luijt RB, van Rooijen E, et al. Genetic analysis of von Hippel-Lindau disease. Hum Mutat 2010;31:521-37. [PubMed]
- Ganeshan D, Menias CO, Pickhardt PJ, et al. Tumors in von Hippel-Lindau Syndrome: From Head to Toe-Comprehensive State-of-the-Art Review. Radiographics 2018;38:849-66. [Crossref] [PubMed]
- McNeill A, Rattenberry E, Barber R, et al. Genotype-phenotype correlations in VHL exon deletions. Am J Med Genet A 2009;149A:2147-51. [Crossref] [PubMed]
- Crona J, Ljungstrom V, Welin S, et al. Bioinformatic Challenges in Clinical Diagnostic Application of Targeted Next Generation Sequencing: Experience from Pheochromocytoma. PLoS One 2015;10:e0133210. [Crossref] [PubMed]
- Gläsker S, Bender BU, Apel TW, et al. The impact of molecular genetic analysis of the VHL gene in patients with haemangioblastomas of the central nervous system. J Neurol Neurosurg Psychiatry 1999;67:758-62. [Crossref] [PubMed]
- Dahia PL. Pheochromocytoma and paraganglioma pathogenesis: learning from genetic heterogeneity. Nat Rev Cancer 2014;14:108-19. [Crossref] [PubMed]
- Knauth K, Bex C, Jemth P, et al. Renal cell carcinoma risk in type 2 von Hippel-Lindau disease correlates with defects in pVHL stability and HIF-1alpha interactions. Oncogene 2006;25:370-7. [Crossref] [PubMed]
- Gnarra JR, Tory K, Weng Y, et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet 1994;7:85-90. [Crossref] [PubMed]


