
Validation of molecular biomarkers for preoperative diagnostics of human papillary thyroid carcinoma in fine needle aspirates
Introduction
Thyroid cancer is the most commonly diagnosed endocrine malignancy with increasing incidence over the last three decades (1). It comprises well-differentiated thyroid carcinomas, including papillary (PTC) and follicular thyroid cancer (FTC), along with poorly differentiated and undifferentiated (anaplastic) thyroid carcinomas which are less common but more aggressive. Current preoperative examination combines ultrasound characteristics with cytological analysis on material obtained from ultrasound-guided fine-needle aspiration (FNA) biopsy. FNA is recommended for the clinical evaluation of most thyroid nodules ≥1 cm (2,3), and represents the test of choice for preoperative diagnosis of thyroid malignancies. The FNA based diagnostics follows the Bethesda reporting system (4), which classifies the sample into one of the categories I–VI, from low (i.e., II—benign: 0–3%) to high malignancy risk (i.e., VI—malignant: 97–99%), based on the cytomorphology. The FNA based diagnostics has the lowest reliability for the Bethesda diagnostic categories bearing intermediate malignancy risk: category III, atypia of undetermined significance (10–30%); category IV, follicular neoplasm or suspicious for a follicular neoplasm (25–40%); and category V, suspicious for malignancy (50–75%) (3,4). Recently, the noninvasive encapsulated follicular variant of PTC, which accounted for 10–25% of all newly diagnosed thyroid cancers in Europe & North America a few years ago, was reclassified into a nonmalignant entity: noninvasive follicular thyroid neoplasm with papillary-like features (NIFTP). NIFTP is now considered as a separate subgroup of thyroid neoplasm, based on specific histopathological features including the absence of vascular or capsular invasion, and is associated with a very low risk of adverse outcomes (4,5). NIFTP reclassification has led to a significant drop in the risk of malignancies for the 3 indeterminate categories (Bethesda III–V) where most of these cases (90%) are diagnosed cytologically. Thus, the value of FNA diagnostics depends on the subtype of thyroid malignancy, being more sensitive for the classical PTC cases, but less sensitive for FTC and follicular variant PTC (FVPTC) including NIFTP. Consequently, surgery is generally recommended in such cases (6), although about 70% of the nodules evaluated preoperatively as indeterminate are classified as benign postoperatively.
In an attempt to increase precision of preoperative diagnostics of indeterminate thyroid nodules, numerous studies identified potential molecular biomarkers with strongly altered expression levels in PTC, including TIMP1 (7), c-KIT (8), CHECK1, c-MET, TPO and BMAL1 (ARNTL) (9-11). This was achieved by comparing side by side the expression profile of large panels of recently identified biomarkers for thyroid malignancies in large-scale studies, including our own works on postoperative formalin-fixed paraffin embedded samples reporting mRNA profiles along with mutation analysis (BRAFV600E) (10,11). Despite these substantial efforts, there are still no reliable preoperative diagnostic markers available for patients with indeterminate thyroid FNA cytology. Therefore, increasing the accuracy of preoperative tests for indeterminate nodules stays of great importance, with the ultimate goal to avoid unjustified surgeries (12,13).
To this aim, we conducted a prospective study, comparing expression of the panel of molecular biomarkers in FNA samples from benign and malignant nodules. Highly sensitive Cell-to-CT and QuantiGene Plex transcript analyses have been performed on the preoperative thyroid FNA samples with available postoperative diagnostics. The biomarker alterations in FTC and PTC FNAs were compared between the two methods and correlated to the previous analyses based on postoperative samples (10,11).
Methods
Study participants and thyroid sampling
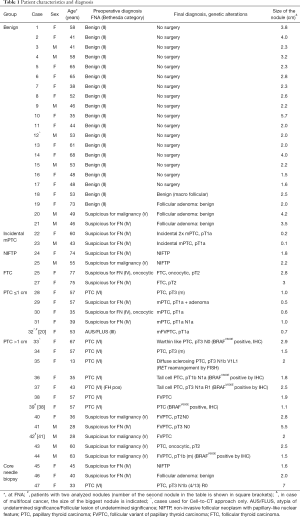
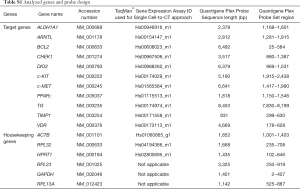
The study enrolled 41 patients who underwent thyroid nodule examination with preoperative ultrasound-guided FNA at the University Hospital of Geneva (Table 1, rows 1–44), which was followed by the surgery in case of pathological findings (Table 1, rows 22–44). The FNA procedure was performed between 2 PM and 6 PM, with written informed consent obtained from each patient. The study protocol was approved by the local Ethics Comity (CCER 14-109). Thyroid cytopathology report was based on the Bethesda reporting system (6). Patient characteristics, FNA based conclusion, and final diagnostics are summarized in Table 1. For QuantiGene Plex assay validation, perioperative core-needle biopsies (CNBs) were performed from three different regions of the same nodule in 3 patients (Table 1, rows 45–47). Postoperative diagnostics of the malignant tumors was done by histopathological analysis according to the World Health Organization Classification of Thyroid Tumors (14) and staged according to the AJCC Cancer Staging Manual 7th ed. For transcriptional analyses, 6 housekeeping genes and 11 potential biomarkers (Table S1) were selected based on our own previous studies (10,11) and on literature search (7,8,15). In terms of classification of cases according to risk of malignancy, invasive FVPTC was considered as a subset of PTC, whereas NIFTP cases, representing very low-risk malignancy group (5,16,17), were analyzed separately from the PTC. Based on the nodule size, the PTC samples were subdivided into papillary thyroid microcarcinomas (<1 cm, mPTC) and PTC (>1 cm).
Full table
Full table
Gene expression quantification by Single Cell-to-CT™ approach
Washout samples from the FNA syringes were diluted in 50 µL of phosphate buffer saline (PBS). 10 µL of the diluted sample was processed with Single Cell-to-CT Kit™ (Ambion/Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. In brief, the assay comprised the following steps: cell lysis, reverse-transcription reaction, c-DNA pre-amplification with 14 specific TaqMan probes (Table S1), and amplification of c-DNA using Real-Time PCR system with the same probes. Amplification step was done in technical duplicates. Target gene expression levels were normalized to the mean of three housekeeping genes (ACTB, HPRT1, RPL13A) using ΔΔCt method, and were log2 transformed for further analysis.
Multiplex gene expression quantification using labeled DNA probes (QuantiGene Plex 2.0 assay)
The QuantiGene Plex 2.0 assay (Thermo Fisher Scientific, Waltham, MA, USA) was used for quantification of target-specific RNAs via the direct hybridization of corresponding branched DNA followed by the amplification of the signal. Target-specific probes premixed with the magnetic Capture Beads (listed in Table S1) were designed by Affymetrix, Thermo Fisher Scientific, Waltham, MA, USA. 20 µL of FNA sample in technical duplicates was directly used as input. The QuantiGene Plex assay was performed according manufacturer’s instructions implying the following steps: cell lysis; pre-incubation of mRNA with probe sets; mixing with the capture beads followed by overnight hybridization; the signal amplification by sequential hybridization to the captured target RNAs via specific Luminex beads. The resulting signal, associated with target-specific capture beads, was detected by the Luminex 200 Flow Cytometer instrument. The QuantiGene Plex signal was expressed as median fluorescence intensity, proportional to the number of target RNA molecules in the samples. Background correction was made by subtracting the mean +2 standard deviation (SD) values from the raw counts obtained with six negative controls. Samples with at least 1 housekeeping gene signal below the detection level were excluded from the analysis. Values less than 1 were fixed to 1 to avoid negative values after log transformation. Counts of genes were then normalized with the geometric mean of 6 housekeeping genes (ACTB, HPRT1, GAPDH, RPL13A, RPL23, and RPL32). Normalized data were then log2 transformed for further analysis.
Statistical analysis
Pair-wise Pearson correlation analysis was applied to test correlation between gene expression data obtained by QuantiGene Plex assay, and between gene expression data obtained with Single Cell-to-CT and QuantiGene Plex approaches for the same patient. Correlation strength was interpreted as described in detail in (11). One-tailed, unpaired Student’s t-test was used to test the significance of difference between PTC and benign groups. A conservative significance threshold of P<0.05 associated with a fold change value ≥2 was applied. The difference between groups within suspicious nodules was tested by one-way ANOVA test with Tukey’s multiple comparison post-test.
Results
Single Cell-to-CT analysis of transcript expression patterns in FNA samples from benign and PTC nodules
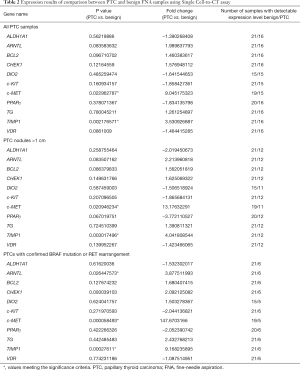
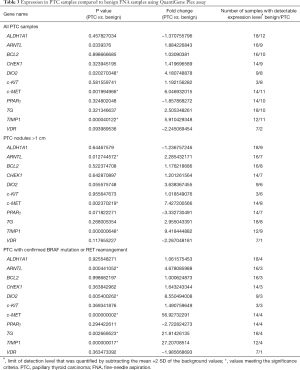
One of the major challenges for reliable molecular analyses of FNA samples is generally low amount of material, along with cellular heterogeneity stemming from the presence of skin, stromal or blood cells. We thus attempted to apply sensitive Single Cell-to-CT assay allowing to measure target gene expression directly on the cell lysates which contain as little as tens to hundreds cells, based on qRT-PCR following the template preamplification step (18). Overall, 44 FNA samples were collected from 41 patients, encompassing 21 benign, 2 NIFTP, 2 FTC, 17 PTC and 2 incidental mPTC samples, according to the final diagnostics done preoperatively for benign, and postoperatively for the rest of samples (Table 1). The panel of 11 target genes (Table S1), previously identified by us as the most predictive of thyroid malignancies based on postoperative sample analyses (10,11), was now analyzed on preoperative FNAs. The genes of interest related to thyroid function, circadian clock, cell cycle and apoptosis were analyzed. One out of 44 samples showed no detectable signal for any gene following amplification (97.7% sensitivity). Most of the target genes showed detectable expression levels in the rest of analyzed samples (Table 2), excluding c-MET (undetectable in 4 samples), PPARγ (undetectable in 1 sample), c-KIT (undetectable in 1 sample), and DIO2 (undetectable in 10 samples). We next compared target gene expression profiles between FNA obtained from benign and confirmed PTC nodules. Statistical analysis revealed significantly higher expression of c-MET and TIMP1 in PTC as compared to benign FNA samples, and similar tendency for circadian core-clock transcript ARNTL, which did not reach statistical significance (Table 2, upper part). The size of the thyroid nodule over 1 cm was previously shown to be associated with higher risk of malignancy (3,19). Thus, we separately analyzed FNA sub-group of PTC nodules bearing the size over 1 cm, after excluding mPTC samples with the nodule size ≤1 cm. In line with the results for all PTC samples together, TIMP1 and c-MET showed significant upregulation in this sub-group (Table 2, middle part). ARNTL was over 2-fold upregulated while PPARγ and ALDH1A1 were over 2-fold downregulated, with none of these changes being statistically significant (Table 2, middle part). Most substantial changes of ARNTL, TIMP1 and c-MET expression were observed in PTC sub-group with confirmed BRAFv600E mutation or RET rearrangement, including tall cell variant and diffuse sclerosing variant [Table 2, lower part; (20,21)]. Moreover, non-significant tendencies for downregulation were observed for PPARγ and c-KIT in this sub-group, while CHECK1 and TG were upregulated (Table 2, lower part).
Full table
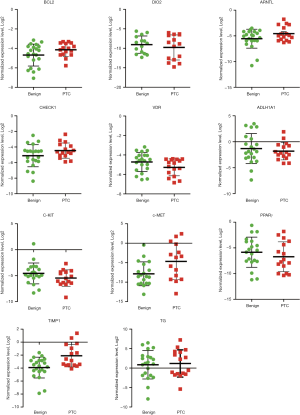
Overall, the alterations in the biomarkers obtained by Single Cell-to-CT analysis in preoperatively collected FNAs entirely corroborated with previously observed tendencies for the same markers measured by NanoString approach in postoperative formalin-fixed paraffin-embedded samples (11), confirming the diagnostic value of these biomarkers. Of note, the variability of each gene expression levels among the samples within the same group was very high (Figure S1), also in the benign group of samples. Such inter-individual variability diminishes diagnostics value of this method.
QuantiGene Plex assay allows for reliable detection of gene expression level in FNA samples
In order to avoid the pre-amplification step used in Cell-to-CT assay, which may lead to certain inaccuracy, we next applied the QuantiGene Plex method based on the direct probe hybridization to the same FNA samples (22-24). In order to validate that the amount of material in FNA is sufficient for reliable transcript detection by this recently developed methodology, a panel of 6 housekeeping genes (Table S1) was first measured in 1 benign and 1 PTC FNA samples (Figure 1). Moreover, we validated the reproducibility of the method by measuring the same housekeeping genes in CNB samples obtained after thyroidectomy from three regions of the same nodule obtained from three patients (Table 1, rows 45–47). The results for all the assessed transcripts demonstrated reliably measurable values in FNA samples, and high reproducibility between 3 replicates of CNB samples (Figure 1).
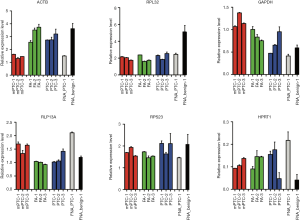
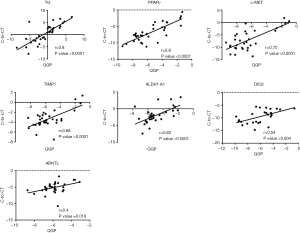
We next analyzed the target gene expression (Table S1) in 41 out of 44 FNA samples (Table 1) by QuantiGene Plex method. The signal for housekeeping genes and most of the target genes were below detection level in 6 out of 41 samples (14.6%). Important inter-individual variations within the groups, similar to those in Cell-to-CT (compare Figures S1 and S2) were observed, rendering a clear separation between PTC and benign clusters difficult in most of the cases. Overall, similar tendencies for alterations of the assessed biomarkers were observed by QuantiGene Plex method (Table 3), as compared to Cell-to-CT results (Table 2). Pearson correlation analysis conducted between the measures obtained by the two methods for each sample/gene suggested very strong positive correlation for TG and PPARγ; strong correlation for c-MET, TIMP1, ALDH1A1, DIO2; and moderate correlation for ARNTL (Figure 2). In contrast, c-KIT, CHECK1, BCL2 and VDR values obtained by the two methods exhibited very weak or weak correlation.
Full table
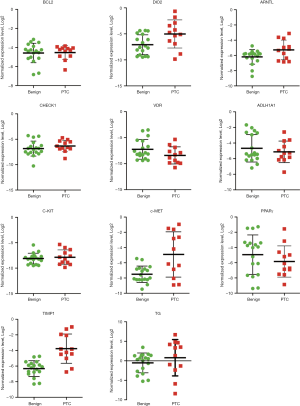
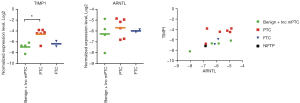
The expression levels of ARNTL, c-MET and TIMP1 measured by QuantiGene Plex were significantly altered in PTCs comprising or not mPTC (Table 3, upper and middle parts). Additionally, 3-fold upregulation of DIO2 was observed in PTCs. In the subset of PTCs with BRAF or RET alteration (Table 3, lower part), ARNTL was upregulated over 4 folds, c-MET over 56 folds, and TIMP1 over 27 folds, all with the high significance levels. PPARγ had 3-fold lower expression in PTC nodules over 1 cm, without reaching significance. Of note, Pearson’s correlation analysis showed strong correlations between ARNTL, c-MET and TIMP expressed in the same samples, which allowed for better separation of PTC and benign sample clusters (Figure 3).

QuantiGene Plex analysis of FNA samples suggests molecular signature for indeterminate nodules
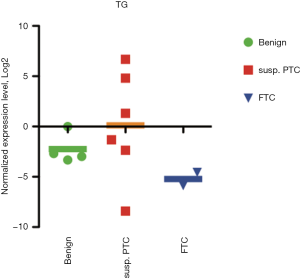
In our sample collection, 15 nodules were classified as indeterminate by cytopathological examination (Bethesda III–V). Within this group, the postoperative diagnostic revealed 2 benign, 2 NIFTP, 2 FTC, 2 incidental mPTC (13%) and 7 PTC cases (48%) (Table 1). Since the incidental mPTC were not the target nodules sampled by FNA, we grouped them with the benign samples for this analysis (Figure 4). One out of two NIFTP and one out of seven PTCs showed undetectable levels of housekeeping genes by QuantiGene Plex, and thus were not taken into account. Of the applied biomarkers, TIMP1 gave the most reliable separation of PTC group from both benign and FTC samples (Figure 4, left panel). Thus, within the group of indeterminate nodules, the sensitivity of TIMP1 overexpression used as predictive biomarker for PTCs with cut-off set at log2 =−5 was 83%, and the specificity was 100%. Although ARNTL expression level was not significantly different between the groups, it exhibited strong correlation with TIMP1 (r=0.66; Figure 4, middle and right panels), separating PTC samples with high accuracy. For FTC samples, the largest alteration was observed for TG expression (over 7-fold decrease), without reaching significance according to ANOVA test (Figure S3), whereas CHECK1, c-KIT, c-MET and DIO2 showed a trend for upregulation (data not shown).
Discussion
Application of gene expression analysis in FNA samples for pre-operative diagnostics of human thyroid follicular malignancies
FNA samples contain vastly variable cell number, ranging from few cells to millions (25). Such variability represents an important challenge for preoperative molecular diagnostics, and for the choice of adequate assay for reliable measurement of gene expression. Beyond the targeted follicular cells from the nodule of interest, FNA samples are frequently contaminated with blood cells, skin and stromal cells, inflammatory cells, and thyrocytes from neighboring healthy or abnormal thyroid tissue (e.g., chronic thyroiditis), which may hinder the conclusions based on the molecular biomarkers expression. Here we conducted a parallel expression analysis of the panel of molecular biomarkers in FNA samples by two recently developed approaches based on (I) amplification of reverse-transcribed cDNA molecules pre-amplified in a target-specific manner (Cell-to-CT), and (II) direct RNA hybridization appropriate for low sample input (QuantiGene Plex). Importantly, overall tendencies for the target gene expression were comparable when assessed by the two approaches (Figures S1,S2; Tables 2,3), validating both approaches for the molecular diagnostics of high precision. Most of target genes values showed average to very strong correlations when measured by two methods (Figure 2). Transcripts with low or no correlation (c-KIT, BCL2, CHEK1 and VDR) should be further tested on higher number of FNA samples in order to validate the most reliable method for their measurement.
The sensitivity of QuantiGene Plex was inferior to the one of Cell-to-CT (85.4% vs. 97.7%), likely due to pre-amplification and amplification steps in the latter. Consequently, the number of target genes, which were below the detection limit, was higher when assessed by QuantiGene Plex as compared to Cell-to-CT (Table 3). Absolute expression levels in the target genes, with the exception of TG, were low (<50 counts, linear scale) in most of the samples when measured by QuantiGene Plex. Despite higher sensitivity, Cell-to-CT assay may result in biased measurements proportional to the high number of pre-amplification cycles, in particular for the low abundant genes (26). Moreover, frequent sample contamination with non-thyrocyte cells taken as an input for pre-amplification may lead to additional bias. High sensitivity of Cell-to-CT kit to the blood cell and even to PBS contamination, along with target gene pre-amplification without a possibility to control the quality of the material at intermediate steps, are disadvantageous for using this type of assay for FNA sample molecular analysis. In addition, different Cell-to-CT kits are calibrated for a very narrow range of the input material each, meaning that distinct types of Cell-to-CT kit should be applied depending on the cell number in each FNA. By contrast, same QuantiGene Plex kit is readily usable for FNA samples containing different amounts of material, and for CNBs (Figure 1). Additional challenge for analyzing gene expression in FNA samples is that due to the sample heterogeneity a classical normalization to housekeeping genes expressed across most of cell types may not be as accurate in this case. On the other hand, application of thyrocyte-specific genes cannot help evaluating amount of thyrocytes within the sample, since most of the thyrocyte-specific genes express altered levels upon the malignancies and cannot be used for normalization. Vast variability of TG expression observed within our FNA sample collection might be explained indeed by the sample heterogeneity (Figures S1,S2).
Finally, morphological and molecular heterogeneity of thyroid nodules should also be considered when relying on molecular diagnostics in FNA samples. It stems from intra-nodular tumor heterogeneity frequently implying different sub-clones of tumor cells (27), also reported for genetic rearrangements in thyroid malignancies (28). Such molecular cell heterogeneity may account for important variability of gene expression even within benign thyroid nodules [Figures S1,S2; (29)]. In rare cases, thyroid carcinoma arises from existing adenomatous nodule (30), resulting in mixed cell population within the nodule. In such situation, the molecular profile resulting from FNA sample analysis might be biased or even exclusively representative for one or another sub-population, leading to erroneous diagnostics. Single cell RNA sequencing of the cells obtained from fresh FNA samples from benign, FTC and PTC nodules might be highly instrumental in order to gather reliable information on the heterogeneity of human thyroid follicular carcinomas.
Potential biomarkers for PTC pre-operative diagnostics
In a good agreement with previously reported biomarkers for PTC diagnostics based on the studies in post-operative material (7,11,31), TIMP1 and c-MET showed the most prominent and highly significant alterations in pre-operative FNA samples between benign and PTC groups (Tables 2,3). Indeed, some of the most aggressive types of PTCs (Diffuse sclerosing, Tall cell) within our cohort showed the largest upregulation in these transcripts, suggesting that these biomarkers are also predictive of the disease progression. TIMP1 transcript upregulation has been associated with BRAF mutations via NF-kB activation. Along with transcriptional data, in 56% of PTC cases TIMP1 protein was identified immunohistochemically, whereas it was undetectable in the benign thyroid tissues (32,33). Likewise, activation of c-MET in cancers often corroborates with poor prognosis and increased drug resistance (34). Interestingly, c-MET and TIMP1 were downregulated in cases of hyperplasia and adenoma, two common benign pathologies of neighboring parathyroid glands which can sometimes mimic a thyroid nodule clinically and cytologically (35).
Consistently with previous molecular studies based on the postoperative samples (11,36), expression of the core-clock gene ARNTL was upregulated in FNA samples obtained pre-operatively from PTCs (Tables 2,3). Alterations in the components of the core-clock machinery have been associated with the wide variety of pathologies, and thus hold a strong potential as predictive biomarkers for these diseases (37-40). Indeed, the observed alterations in ARNTL expression in PTCs corroborate with progressive changes of the molecular clocks that were proposed to be linked to the human thyroid cancer progression (36,41). Because the causality of this connection stays largely unexplored, further studies in human primary thyrocytes established from PTC nodules will be required to scrutinize the role of ARNTL and of the molecular clock in general in human thyroid cancer progression. Importantly, strong positive correlation between c-MET, TIMP1 and ARNTL observed by us in FNA samples (Figures 3,4) goes well along with our observations in postoperative samples (11). The observed correlation may suggest functional interactions between these gene products during the progression of thyroid carcinomas. The connection between the core-clock component ARNTL with TIMP1 and c-MET needs to be further explored, since it holds promise for the preoperative diagnostic purposes, and might provide novel insights into our understanding of thyroid carcinomas etiology.
FTC, FVPTC and NIFTP represent the most difficult challenge for pre-operative FNA based diagnostics, and account for most malignancies identified in cytologically indeterminate nodules. Although TG may hold promise for FTC diagnostics (Figure 4), consistently with our previous analysis of post-operative FTCs (10), a very low number of these samples in our current collection renders statistically significant conclusions impossible. Further analyses of these biomarkers in FNA by QuantiGene Plex will be required.
In addition to the gene expression analysis, pre-operative molecular diagnostics may include gene mutations/rearrangements. Recently developed successful analyses of gene mutations/rearrangements in FNA samples predictive of FTCs and PTCs (13,42,43), combined with biomarker expression profiling of mRNA or miRNAs, may increase the preoperative diagnostics precision (44). CNB sampling has been raised as an alternative to FNA for the preoperative diagnostics in cases with previous indeterminate cytology (Bethesda III–V) or unsatisfactory material (Bethesda I) (45-47). While CNB is significantly more invasive as compared to FNA sampling, it allows obtaining considerably more material, including both lesional and non-lesional tissues, which will likely result in reliable molecular testing and mutation analysis (Figure 1) (48,49). Finally, recently developed approach for cancer diagnostics based on the tissue-specific methylation profile of mutant DNA measurement in the blood samples (50) may represent a powerful approach for non-invasive and reliable diagnostics of thyroid malignancies (13).
Acknowledgments
This work was funded by the Fondation pour la Recherche sur le Cancer et la Biologie and Fonds de Recherche du Département des Spécialités de Médecine (CD).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study protocol was approved by the local Ethics Comity (CCER 15-109). Written informed consent obtained from each patient.
References
- Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin 2014;64:9-29. [Crossref] [PubMed]
- Alexander EK, Kennedy GC, Baloch ZW, et al. Preoperative diagnosis of benign thyroid nodules with indeterminate cytology. N Engl J Med 2012;367:705-15. [Crossref] [PubMed]
- Haugen BR. 2015 American Thyroid Association Management Guidelines for Adult Patients with Thyroid Nodules and Differentiated Thyroid Cancer: What is new and what has changed? Cancer 2017;123:372-81. [Crossref] [PubMed]
- Cibas ES, Ali SZ. The 2017 Bethesda System for Reporting Thyroid Cytopathology. Thyroid 2017;27:1341-6. [Crossref] [PubMed]
- Nikiforov YE, Seethala RR, Tallini G, et al. Nomenclature Revision for Encapsulated Follicular Variant of Papillary Thyroid Carcinoma: A Paradigm Shift to Reduce Overtreatment of Indolent Tumors. JAMA Oncol 2016;2:1023-9. [Crossref] [PubMed]
- Cibas ES, Ali SZ. The Bethesda System for Reporting Thyroid Cytopathology. Thyroid 2009;19:1159-65. [Crossref] [PubMed]
- Ilie MI, Lassalle S, Long-Mira E, et al. In papillary thyroid carcinoma, TIMP-1 expression correlates with BRAF (V600E) mutation status and together with hypoxia-related proteins predicts aggressive behavior. Virchows Archiv 2013;463:437-44. [Crossref] [PubMed]
- Tomei S, Mazzanti C, Marchetti I, et al. c-KIT receptor expression is strictly associated with the biological behaviour of thyroid nodules. J Transl Med 2012;10:7. [Crossref] [PubMed]
- Romei C, Ciampi R, Faviana P, et al. BRAFV600E mutation, but not RET/PTC rearrangements, is correlated with a lower expression of both thyroperoxidase and sodium iodide symporter genes in papillary thyroid cancer. Endocrine-related cancer 2008;15:511-20. [Crossref] [PubMed]
- Makhlouf AM, Chitikova Z, Pusztaszeri M, et al. Identification of CHEK1, SLC26A4, c-KIT, TPO and TG as new biomarkers for human follicular thyroid carcinoma. Oncotarget 2016;7:45776-88. [Crossref] [PubMed]
- Chitikova Z, Pusztaszeri M, Makhlouf AM, et al. Identification of new biomarkers for human papillary thyroid carcinoma employing NanoString analysis. Oncotarget 2015;6:10978-93. [Crossref] [PubMed]
- Vaccarella S, Franceschi S, Bray F, et al. Worldwide Thyroid-Cancer Epidemic? The Increasing Impact of Overdiagnosis. N Engl J Med 2016;375:614-7. [Crossref] [PubMed]
- Dibner C, Sadowski SM, Triponez F, et al. The search for preoperative biomarkers for thyroid carcinoma: application of the thyroid circadian clock properties. Biomark Med 2017;11:285-93. [Crossref] [PubMed]
- The International Agency for Research on Cancer. In: Lloyd R, Heitz P, Eng C. editors. WHO classification of tumours of endocrine organs. 4th edition. Lyon: IARC Press, 2017.
- Hawthorn L, Stein L, Varma R, et al. TIMP1 and SERPIN-A overexpression and TFF3 and CRABP1 underexpression as biomarkers for papillary thyroid carcinoma. Head Neck 2004;26:1069-83. [Crossref] [PubMed]
- Johnson DN, Cavallo AB, Uraizee I, et al. A Proposal for Separation of Nuclear Atypia and Architectural Atypia in Bethesda Category III (AUS/FLUS) Based on Differing Rates of Thyroid Malignancy. Am J Clin Pathol 2019;151:86-94. [PubMed]
- Díaz Del Arco C, Fernandez Acenero MJ. Noninvasive Follicular Thyroid Neoplasm with Papillary-Like Nuclear Features: Can Cytology Face the Challenge of Diagnosis in the Light of the New Classification? Acta Cytol 2018;62:265-72. [Crossref] [PubMed]
- Luger R, Valookaran S, Knapp N, et al. Toll-like receptor 4 engagement drives differentiation of human and murine dendritic cells from a pro- into an anti-inflammatory mode. PLoS One 2013;8:e54879. [Crossref] [PubMed]
- Russ G, Bonnema SJ, Erdogan MF, et al. European Thyroid Association Guidelines for Ultrasound Malignancy Risk Stratification of Thyroid Nodules in Adults: The EU-TIRADS. Eur Thyroid J 2017;6:225-37. [Crossref] [PubMed]
- Lee MY, Ku BM, Kim HS, et al. Genetic Alterations and Their Clinical Implications in High-Recurrence Risk Papillary Thyroid Cancer. Cancer Res Treat 2017;49:906-14. [Crossref] [PubMed]
- Kazaure HS, Roman SA, Sosa JA. Aggressive variants of papillary thyroid cancer: incidence, characteristics and predictors of survival among 43,738 patients. Ann Surg Oncol 2012;19:1874-80. [Crossref] [PubMed]
- Hall JS, Usher S, Byers RJ, et al. QuantiGene Plex Represents a Promising Diagnostic Tool for Cell-of-Origin Subtyping of Diffuse Large B-Cell Lymphoma. J Mol Diagn 2015;17:402-11. [Crossref] [PubMed]
- Tian Y, Pan F, Sun X, et al. Association of TET1 expression with colorectal cancer progression. Scand J Gastroenterol 2017;52:312-20. [Crossref] [PubMed]
- Chinzei N, Rai MF, Hashimoto S, et al. Evidence for Genetic Contribution to Variation in Post-Traumatic Osteoarthritis in Mice. Arthritis Rheumatol 2019;71:370-81. [PubMed]
- Alshaikh S, Harb Z, Aljufairi E, et al. Classification of thyroid fine-needle aspiration cytology into Bethesda categories: An institutional experience and review of the literature. Cytojournal 2018;15:4. [Crossref] [PubMed]
- Korenková V, Scott J, Novosadova V, et al. Pre-amplification in the context of high-throughput qPCR gene expression experiment. BMC Mol Biol 2015;16:5. [Crossref] [PubMed]
- Almendro V, Marusyk A, Polyak K. Cellular heterogeneity and molecular evolution in cancer. Annu Rev Pathol 2013;8:277-302. [Crossref] [PubMed]
- Finkel A, Liba L, Simon E, et al. Subclonality for BRAF Mutation in Papillary Thyroid Carcinoma Is Associated With Earlier Disease Stage. J Clin Endocrinol Metab 2016;101:1407-13. [Crossref] [PubMed]
- Pagni F, Jaconi M, Delitala A, et al. Incidental papillary thyroid carcinoma: diagnostic findings in a series of 287 carcinomas. Endocr Pathol 2014;25:288-96. [Crossref] [PubMed]
- Fusco A, Chiappetta G, Hui P, et al. Assessment of RET/PTC oncogene activation and clonality in thyroid nodules with incomplete morphological evidence of papillary carcinoma: a search for the early precursors of papillary cancer. Am J Pathol 2002;160:2157-67. [Crossref] [PubMed]
- Sierra JR, Tsao MS. c-MET as a potential therapeutic target and biomarker in cancer. Ther Adv Med Oncol 2011;3:S21-35. [Crossref] [PubMed]
- Wasenius VM, Hemmer S, Kettunen E, et al. Hepatocyte growth factor receptor, matrix metalloproteinase-11, tissue inhibitor of metalloproteinase-1, and fibronectin are up-regulated in papillary thyroid carcinoma: a cDNA and tissue microarray study. Clin Cancer Res 2003;9:68-75. [PubMed]
- Bommarito A, Richiusa P, Carissimi E, et al. BRAFV600E mutation, TIMP-1 upregulation, and NF-kappaB activation: closing the loop on the papillary thyroid cancer trilogy. Endocr Relat Cancer 2011;18:669-85. [Crossref] [PubMed]
- Maroun CR, Rowlands T. The Met receptor tyrosine kinase: a key player in oncogenesis and drug resistance. Pharmacol Ther 2014;142:316-38. [Crossref] [PubMed]
- Sadowski SM, Pusztaszeri M, Brulhart-Meynet MC, et al. Identification of Differential Transcriptional Patterns in Primary and Secondary Hyperparathyroidism. J Clin Endocrinol Metab 2018;103:2189-98. [Crossref] [PubMed]
- Mannic T, Meyer P, Triponez F, et al. Circadian clock characteristics are altered in human thyroid malignant nodules. J Clin Endocrinol Metab 2013;98:4446-56. [Crossref] [PubMed]
- Saini C, Brown SA, Dibner C. Human peripheral clocks: applications for studying circadian phenotypes in physiology and pathophysiology. Front Neurol 2015;6:95. [Crossref] [PubMed]
- Gonzalez Rodriguez E, Hernandez A, Dibner C, et al. Arterial blood pressure circadian rhythm: significance and clinical implications. Rev Med Suisse 2012;8:1709. [PubMed]
- Ditisheim AJ, Dibner C, Philippe J, et al. Biological rhythms and preeclampsia. Front Endocrinol (Lausanne) 2013;4:47. [Crossref] [PubMed]
- Dibner C, Schibler U. Circadian timing of metabolism in animal models and humans. J Intern Med 2015;277:513-27. [Crossref] [PubMed]
- Philippe J, Dibner C. Thyroid circadian timing: roles in physiology and thyroid malignancies. J Biol Rhythms 2015;30:76-83. [Crossref] [PubMed]
- Ferrari SM, Fallahi P, Ruffilli I, et al. Molecular testing in the diagnosis of differentiated thyroid carcinomas. Gland Surg 2018;7:S19-S29. [Crossref] [PubMed]
- Decaussin-Petrucci M, Descotes F, Depaepe L, et al. Molecular testing of BRAF, RAS and TERT on thyroid FNAs with indeterminate cytology improves diagnostic accuracy. Cytopathology 2017;28:482-7. [Crossref] [PubMed]
- Nicholson KJ, Yip L. An update on the status of molecular testing for the indeterminate thyroid nodule and risk stratification of differentiated thyroid cancer. Curr Opin Oncol 2018;30:8-15. [Crossref] [PubMed]
- Suh CH, Baek JH, Lee JH, et al. The Role of Core-Needle Biopsy as a First-Line Diagnostic Tool for Initially Detected Thyroid Nodules. Thyroid 2016;26:395-403. [Crossref] [PubMed]
- Suh CH, Baek JH, Lee JH, et al. The role of core-needle biopsy in the diagnosis of thyroid malignancy in 4580 patients with 4746 thyroid nodules: a systematic review and meta-analysis. Endocrine 2016;54:315-28. [Crossref] [PubMed]
- Choi YJ, Baek JH, Suh CH, et al. Core-needle biopsy versus repeat fine-needle aspiration for thyroid nodules initially read as atypia/follicular lesion of undetermined significance. Head Neck 2017;39:361-9. [Crossref] [PubMed]
- Yip L, Nikiforova MN, Carty SE, et al. Optimizing surgical treatment of papillary thyroid carcinoma associated with BRAF mutation. Surgery 2009;146:1215-23. [Crossref] [PubMed]
- Na DG, Kim JH, Sung JY, et al. Core-needle biopsy is more useful than repeat fine-needle aspiration in thyroid nodules read as nondiagnostic or atypia of undetermined significance by the Bethesda system for reporting thyroid cytopathology. Thyroid 2012;22:468-75. [Crossref] [PubMed]
- Dor Y, Cedar H. Principles of DNA methylation and their implications for biology and medicine. Lancet 2018;392:777-86. [Crossref] [PubMed]


