The role of noncoding RNA in hepatocellular carcinoma
Introduction
Less than 2% the mammalian genome can code protein what called protein coding genes and over 90% of genome represent noncoding RNA (ncRNA) which are transcribed but do not encode proteins (1). In general, ncRNA can be divided into two classes based on their length, short/small ncRNA and long ncRNA. The short ncRNA includes transcripts such as miRNAs, transfer RNAs (tRNAs), small interfering RNAs (siRNAs), piwi-interacting RNAs (piRNAs) and some ribosomal RNAs. Previous studies have revealed that these short ncRNAs play important roles in many biological processes (2). Although comparatively less understood in human cancers, increasing studies indicated long ncRNAs play crucial role in regulating numerous development and biological pathway in genome (3,4). As the one of most common cancers worldwide, primary hepatocellular carcinoma (HCC) is threatening many people’s health especially in developing countries. The major risk factors for HCC include chronic infections with the hepatitis B or C viruses, heavy drinking, and spoilt foodstuffs contaminated with aflatoxins (5). Among those, the hepatitis B virus (HBV) infection play the critical role in the pathogenesis of HCC for its coherent distribution with the HCC prevalence (6). The present review highlighted the roles of MiRNAs and newly discovered lncRNAs in HCC development, tumorigenesis, metastasis and clinical implication.
MiRNAs and HCC
MicroRNAs (miRNAs) are small non-coding RNAs that may regulate genes expression, either by inhibiting target mRNA translation or inducing its degradation by pairing with complementary sequences within the 3'-untranslated regions (UTRs) of targeted transcripts at the post-transcriptional and/or translational level (7-9) Alterations of miRNA, including expression disorders and mutations, are involved in the initiation and progression of human cancers (10). Increasing evidence has revealed that miRNAs are negative regulators of gene expression and can function as tumor suppressors or oncogenes (11). Among the deregulated miRNAs in HBV-related HCC, some are up-regulated and others down-regulated. Consequently, these molecular changes favor tumor cell proliferation, invasion and metastasis (12). MiRNA expression profiling of human cancer has identified signatures associated with diagnosis prognosis, staging, and response to treatment (13,14). MiRNAs in serum or plasma called circulating miRNAs have been considered as promising noninvasive novel biomarkers for cancer diagnosis and prognosis (15,16). Additionally Yang et al. (17) suggest that urinary miRNA might be a more specific and consistent biomarker of chemical-induced liver injury than the current serum ALT and AST. Accumulating data revealed a subset of miRNAs deregulated in HCC and HBV persistent infection. Out of numerous differentially expressed miRNAs in HCC and HBV persistent infection, miR-122, miR-221, miR-520 were extensively studies. Studies have demonstrated the up-regulation of miR-221/222 and down-regulation of miR-520 and miR-122 in HCC. Liu et al. (18) uncovered that increased risk for HCC in HBV persistent carriers due to the up-regulation of the miR-106b-25 cluster expression. Further study showed that miR-221/222 could enhance cell growth in vitro via targeting CDK inhibitor p27. Conversely, these activities can be efficiently inhibited by an antagomiR specific for miR-221 (11). Significant down-regulation of miR-520 was detected in HCC tissues compared with normal liver cell or adjacent noncancerous liver tissues. By using DIANA microT v3.0 algorithm, TargetScan and PicTar, Zhang w et al. (19) validated that MEKK2 and cyclin D1 were the direct target genes of miR-520b what can lead to abnormal cellular proliferation or development of cancer (20). Pkm2, which is commonly enriched expressed in human embryonic stem cell (hESCs) and HCCs and facilitates self-renewal and proliferation, can be suppressed by miR-122 at translation level (21). Overexpression of miR-122 can lead to a common deficiency in self-renewal and proliferation. Similar inverse relationship between miR-122 and Pkm2 is reflected in the differentiation process of hESCs into hepatocytes. These findings indicate that miRNA-122 possibly serves as a modulator of self-renewal during hepatic lineage specification. Many studies have demonstrated significant difference of miRNA expression profiles in HCC compared with normal or nontumor tissues. Additional studies showed the existence of a large amount of stable miRNAs in routinely collected, formalin-fixed paraffin-embedded (FFPE) samples and in human serum/plasma (22,23). Furthermore, several studies showed that there is no significant deterioration occurring in expression between FFPE and matched fresh-frozen tissues (24,25). The ability to detect miRNA profiles in FFPE tissues or in human serum/plasma implicated a great opportunity to perform the large retrospective analyses needed to confirm the diagnostic and prognostic significance. Serum AFP has been used as a conventional tumor marker for HCC screening long time. AFP <20 ng/mL is considered as normal and AFP >400 ng/mL as true positive in general. The European Society for Medical Oncology (EMSO) guidelines also recommended that elevation of AFP >400 ng/mL can be used instead of fine-needle cytology for diagnosis, especially in patients with liver cirrhosis (26). Despite that, the sensitivity of AFP is just modest (sensitivity: 39-65% and specificity: 76-94%), leaving approximate one-third of early-stage HCC with small tumors (<3 cm) undiagnosed (27). Additionally, serum AFP level is elevated in benign liver diseases, such as hepatitis and cirrhosis. Thus, investigation of miRNA expression pattern could outperform AFP in terms of detecting early-stage HCC, AFP-negative HCC as well as prognostic indicator. Liu et al. (28) investigated expression pattern of miRNAs in resected tumor/adjacent nontumor tissues by real-time quantitative reverse transcription-PCR and revealed that miR-15b,miR-21,miR-130b and miR-183 were highly expressed in both tumor and serum samples. Remarkably, these Serum miRNAs were markedly reduced after surgery, indicating the tumor-derived source of these circulating miRNAs. In addition, when combined miR-15b and miR-130b as a classifier for HCC detection, the detection sensitivity of the classifier in a subgroup of HCCs with low AFP (<20 ng/mL) was 96.7%. The classifier also identified early-stage HCC that could not be detected by AFP. These findings suggested that specific miRNAs could be serum biomarker with clinical implication in both early diagnosis and tumor recurrence after resection.
LncRNAs and HCC
LncRNAs, mRNA-like transcript, are longer than 200 nucleotides but cannot code protein products. Many identified lncRNAs are transcribed by RNA polymerase II (RNA pol II).They can be identified through bioinformatics prediction or a high-throughput analysis such as microarrays and transcriptome analysis (29). Increasing studies have indicated that lncRNAs are crucial regulators of numerous development- and biology-related pathways in genome. Some researchers attempted to classify lncRNAs based on their genomic proximity to protein-coding genes, including five types: (I) sense, (II) antisense, (III) bidirectional, (IV) intronic, and (V) intergenic (Figure 1) (30).
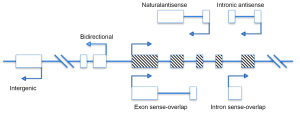
Many studies have indicated that lncRNA transcripts play crucial roles in a variety of biological processes function in Gene imprinting, cell fate specification, cell apoptosis and cell cycle control, Chromatin modification or act as RNA processing Enhancer and promoter associated lncRNAs, nuclear architecture subnuclear compartments (31). LncRNAs utilize varied mechanism in biological processes. In general, the role of lncRNA gene expressing regulator could be found in transcriptional level or post-transcriptional level. Cis- and trans-regulation are two main transcriptional regulations, under which lncRNAs can target genomically local and distant genes respectively. The post- transcriptional regulatory mechanism is involved in post- transcriptional processing of mRNAs, including splicing, editing, trafficking, translation and degradation or function as competing endogenous RNA (ceRNA) for shared miRNA regulating miRNA distribution on specific targets (1).
Out of numerous lncRNAs, a few novel transcripts such as MALAT1, HOTAIR, PRNCR1, PCTAs, H19, HULT are frequently aberrantly expressed in a variety of human cancers. MALAT1 is widely expressed in both normal human and mouse tissues. It shows abnormal expression in numerous human carcinomas, including liver, pancreas, colon, lung, breast, prostate and ovarian cancer (32). RNA interference-mediated silencing of MALAT1 impairs the migration of lung adenocarcinoma cells through regulation of motility-associating genes via transcriptional and/or post-transcriptional level means. Similarly, short hairpin RNA inhibition of MALAT1 reduces cell proliferation, and invasive potential of a cervical cancer cell line (33). Lai et al. (32) suggest that MALAT1 could be a novel biomarker for predicting tumor recurrence after liver transplantation and sever as a promising therapeutic target. Hox transcript antisense intergenic RNA (HOTAIR) is a long ncRNA that trimethylates histone H3 lysine-27 (H3K27me3) of the HOXD locus with the polycomb-repressive complex 2 (PRC2), which is composed of EZH2, SUZ12, and EED, and inhibits HOXD gene expression (34). Thus, HOTAIR epigenetically regulates HOXD expression, located on a different chromosome. Ishibashi et al. (35) reported that Patients with HOTAIR expression had significantly poorer prognoses and a larger primary tumor size than those without HOTAIR expression. Similar to studies in breast and colorectal cancers, H19 is reported to be reactivated during adult tissue regeneration and tumorigenesis. H19 is highly expressed in liver metastasis derived from a range of carcinomas. The H19 gene behaves as an oncogene and may serve as a potential new target for antitumor therapy. LncRNA-HEIH is up regulated in HCC. It plays a key role in the G0/G1 arrest, and is associated with enhancer of zeste homolog 2 (EZH2), which is required for the repression of EZH2 target genes (36). The product of the MYC oncogene is widely deregulated in cancer and functions as a regulator of gene transcription. c-Myc significantly induces the expression of the H19 noncoding RNA in diverse cell types. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis (31). HULC, short for Highly Up regulated in Liver Cancer, is about 1.6 k nucleotide long, containing two exons but not translated to protein. It has been identified that HULC is highly upregulated in HCC and colorectal cancer that metastasized to livers. Liu et al. (37) reported that single nucleotide polymorphisms (SNPs) rs7763881 in HULC was significantly associated with HCC susceptibility in HBV persistent carriers, while rs619586 was protective for non-drinkers’ HCC risk in subgroup analysis. MALAT1 rs619586 was associated with a decreased HCC risk with a borderline significance and was significantly protective for HCC risk in never-drinkers.
Up to now a great number of important details that remain unsolved despite recent progress in lncRNA research. The implication of lncRNAs in tumorigenesis, metastasis, and progression remain to be further investigated. It is to be hoped that the recent burst of interest in lncRNAs will foreshadow resolution of all of these issues from wide-ranging experimental discoveries about the evolution and functional mechanisms of lncRNAs.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- Li X, Wang Z. The role of noncoding RNA in thyroid cancer. Gland Surg 2012;1:146-50.
- ENCODE Project Consortium, Birney E, Stamatoyannopoulos JA, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007;447:799-816.
- Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464:1071-6.
- Yu W, Gius D, Onyango P, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature 2008;451:202-6.
- Varnholt H, Drebber U, Schulze F, et al. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology 2008;47:1223-32.
- Chuang SC, La Vecchia C, Boffetta P. Liver cancer: descriptive epidemiology and risk factors other than HBV and HCV infection. Cancer Lett 2009;286:9-14.
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120:15-20.
- Shukla GC, Singh J, Barik S. MicroRNAs: Processing, Maturation, Target Recognition and Regulatory Functions. Mol Cell Pharmacol 2011;3:83-92.
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215-33.
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006;6:857-66.
- Pineau P, Volinia S, McJunkin K, et al. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A 2010;107:264-9.
- Garzon R, Fabbri M, Cimmino A, et al. MicroRNA expression and function in cancer. Trends Mol Med 2006;12:580-7.
- Sempere LF. Integrating contextual miRNA and protein signatures for diagnostic and treatment decisions in cancer. Expert Rev Mol Diagn 2011;11:813-27.
- Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature 2005;435:834-8.
- Cortez MA, Calin GA. MicroRNA identification in plasma and serum: a new tool to diagnose and monitor diseases. Expert Opin Biol Ther 2009;9:703-711.
- Xu J, Wu C, Che X, et al. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog 2011;50:136-42.
- Yang X, Greenhaw J, Shi Q, et al. Identification of urinary microRNA profiles in rats that may diagnose hepatotoxicity. Toxicol Sci 2012;125:335-44.
- Liu Y, Zhang Y, Wen J, et al. A genetic variant in the promoter region of miR-106b-25 cluster and risk of HBV infection and hepatocellular carcinoma. PLoS One 2012;7:e32230.
- Zhang W, Kong G, Zhang J, et al. MicroRNA-520b inhibits growth of hepatoma cells by targeting MEKK2 and cyclin D1. PLoS One 2012;7:e31450.
- Hayakawa Y, Hirata Y, Nakagawa H, et al. Apoptosis signal-regulating kinase 1 and cyclin D1 compose a positive feedback loop contributing to tumor growth in gastric cancer. Proc Natl Acad Sci U S A 2011;108:780-5.
- Jung CJ, Iyengar S, Blahnik KR, et al. Epigenetic modulation of miR-122 facilitates human embryonic stem cell self-renewal and hepatocellular carcinoma proliferation. PLoS One 2011;6:e27740.
- Zhou J, Yu L, Gao X, et al. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol 2011;29:4781-8.
- Klopfleisch R, Weiss AT, Gruber AD. Excavation of a buried treasure--DNA, mRNA, miRNA and protein analysis in formalin fixed, paraffin embedded tissues. Histol Histopathol 2011;26:797-810.
- Culpin RE, Sieniawski M, Proctor SJ, et al. MicroRNAs are suitable for assessment as biomarkers from formalin-fixed paraffin-embedded tissue, and miR-24 represents an appropriate reference microRNA for diffuse large B-cell lymphoma studies. J Clin Pathol 2013;66:249-52.
- Liu A, Xu X. MicroRNA isolation from formalin-fixed, paraffin-embedded tissues. Methods Mol Biol 2011;724:259-67.
- Verslype C, Rosmorduc O, Rougier P, et al. Hepatocellular carcinoma: ESMO-ESDO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2012;23:vii41-8.
- Collier J, Sherman M. Screening for hepatocellular carcinoma. Hepatology 1998;27:273-8.
- Liu AM, Yao TJ, Wang W, et al. Circulating miR-15b and miR-130b in serum as potential markers for detecting hepatocellular carcinoma: a retrospective cohort study. BMJ Open 2012;2:e000825.
- Yan B, Wang ZH, Guo JT. The research strategies for probing the function of long noncoding RNAs. Genomics 2012;99:76-80.
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009;136:629-41.
- Yan B, Wang Z. Long noncoding RNA: its physiological and pathological roles. DNA Cell Biol 2012;31:S34-41.
- Lai MC, Yang Z, Zhou L, et al. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol 2012;29:1810-6.
- Guo F, Li Y, Liu Y, et al. Inhibition of metastasis-associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta Biochim Biophys Sin (Shanghai) 2010;42:224-9.
- Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007;129:1311-23.
- Ishibashi M, Kogo R, Shibata K, et al. Clinical significance of the expression of long non-coding RNA HOTAIR in primary hepatocellular carcinoma. Oncol Rep 2013;29:946-50.
- Yang F, Zhang L, Huo XS, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology 2011;54:1679-89.
- Liu Y, Pan S, Liu L, et al. A genetic variant in long non-coding RNA HULC contributes to risk of HBV-related hepatocellular carcinoma in a Chinese population. PLoS One 2012;7:e35145.


